Summary
The molecular cloning method involves creating identical copies of a given DNA sequence or gene using a vector and a host organism. It makes perfect duplicates of a gene or DNA sequence (Sakhel et al., 2021). It includes the replication and production of numerous copies of the inserted DNA utilizing a vector, a DNA molecule capable of transporting foreign DNA, and a host organism. The procedure entails extracting the target DNA sequence, cloning it, and introducing the recombinant vector into the host organism. It also involves selecting and screening transformed cells and confirming the presence of the target DNA. Molecular cloning has transformed biological research to analyze and change DNA sequences, comprehend gene function, and create novel biotech and medical applications. This approach can be utilized for various molecular biology processes, including gene expression, genetic engineering, and genetic analysis. Also, when the recombinant vector is introduced into host cells, which produces the therapeutic protein using the cloned DNA as a template. The protein is extracted, purified, quality-checked, and formulated for medical use. This efficient process enables the large-scale production of valuable therapeutic proteins like insulin, growth factors, and antibodies. In addition, the Taq polymerase gene has been cloned and expressed in recombinant hosts like Escherichia coli to create vast amounts of the enzyme. Large amounts of Taq polymerase are required because it is an essential enzyme used in PCR, and the technique involves multiple cycles of amplification, necessitating a sufficient supply of the enzyme to complete each cycle effectively. Large amounts of these proteins may be used in medicines by encoding the genes into appropriate expression vectors and expressing them in host organisms like bacteria, yeast, or mammalian cells to enable synthesis of the required protein. The use of GST fusion proteins is one such application. When a protein of interest is fused with the GST protein, fusion proteins can be employed for protein purification, protein-protein interaction research, and antibody production because of its high affinity for glutathione, the GST protein is useful for protein purification. The immobilized fusion protein can capture additional proteins to study proteinprotein interactions (Simmet et al., 2020). Finally, antibodies for several applications can be generated by employing the GST fusion protein as an antigen. The GST protein is an excellent fusion partner due to its strong affinity for glutathione and ease of separation from the protein of interest.
Method
The Polymerase Chain Reaction (PCR) is a robust molecular biology method for amplifying specific DNA sequences. To execute GST DNA PCR amplification, a reaction mixture of 10 ng of template DNA, 1 μM of each primer, 0.2 mM dNTPs, 2mM MgCl2, 1X Taq buffer, and 0.5 U Taq DNA polymerase was prepared in a total volume of 50 l. The reaction mixture was heated to 95°C for 5 minutes to denature the template DNA, followed by 30 cycles of PCR using the following conditions: 95°C for 30 seconds (denaturation), 55°C for 30 seconds (annealing), and 72°C for 1 minute (extension) (Wang et al., 2020). After 30 cycles, a final extension step at 72°C for 5 minutes was performed to ensure the DNA strands have fully expanded. Electrophoresis was performed using a 1% agarose gel to examine the PCR result. One gram of agarose was dissolved in 100 ml of 1X TAE buffer by heating the mixture until all of the agarose was dissolved. The gel was poured into a gel casting tray after cooling at 60°C. After the gel had been set up, wells are made in the gel using a comb so that the PCR product may be loaded. Using a pipette, the PCR product is combined in a 1:5 ratio with a 6X loading buffer and deposited onto the agarose gel (Wang et al., 2020). The gel was run for 45 minutes at 100 V to separate the DNA fragments according to size.
Results
Discussion
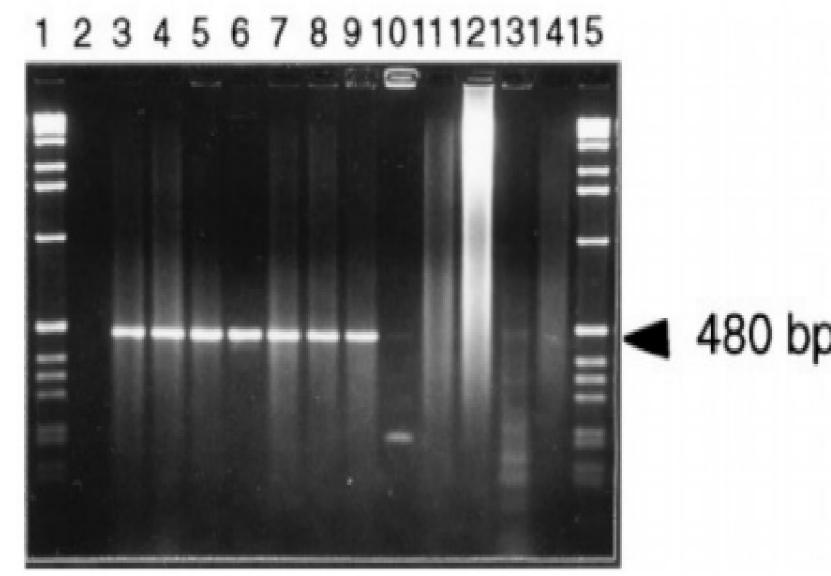
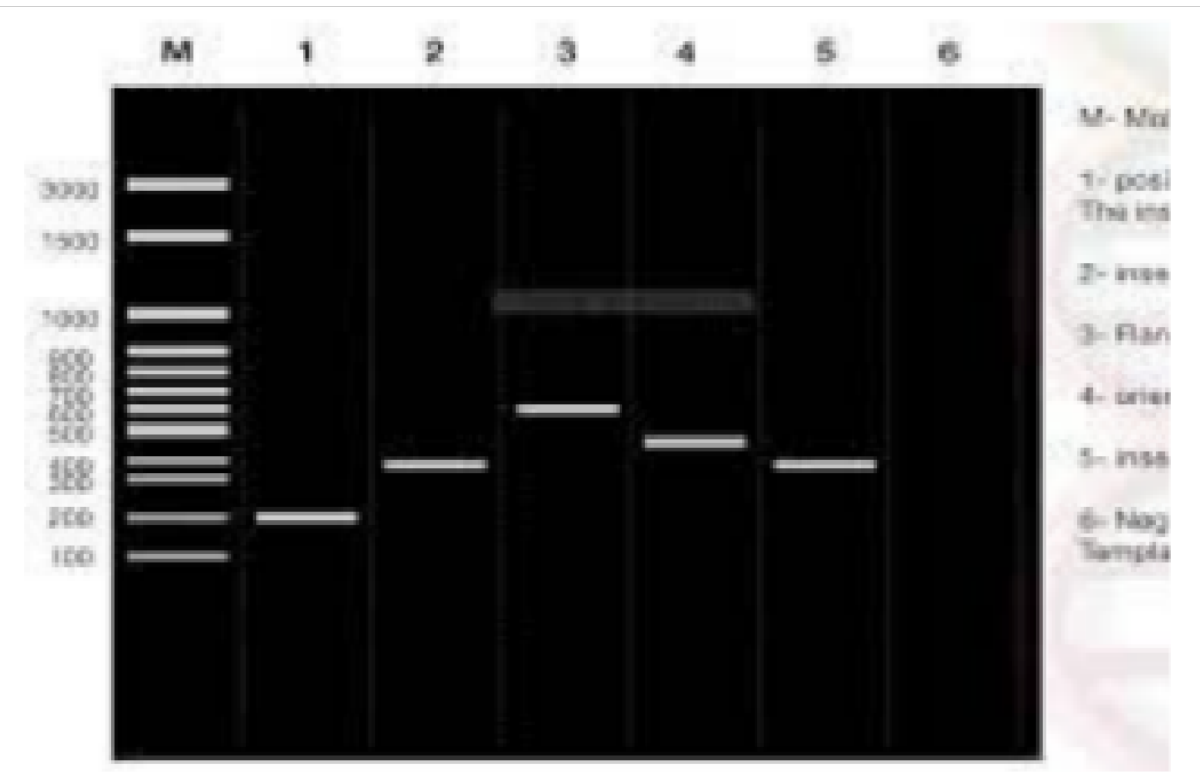
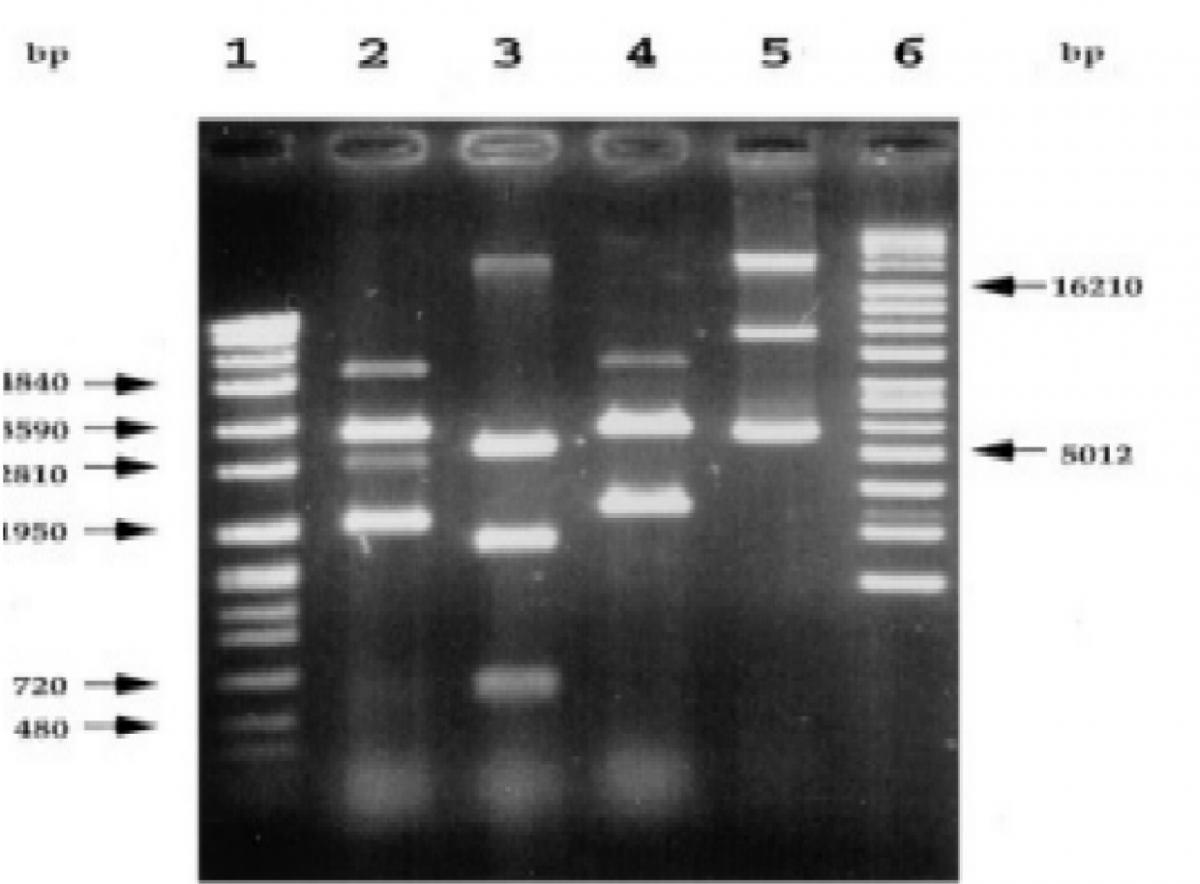
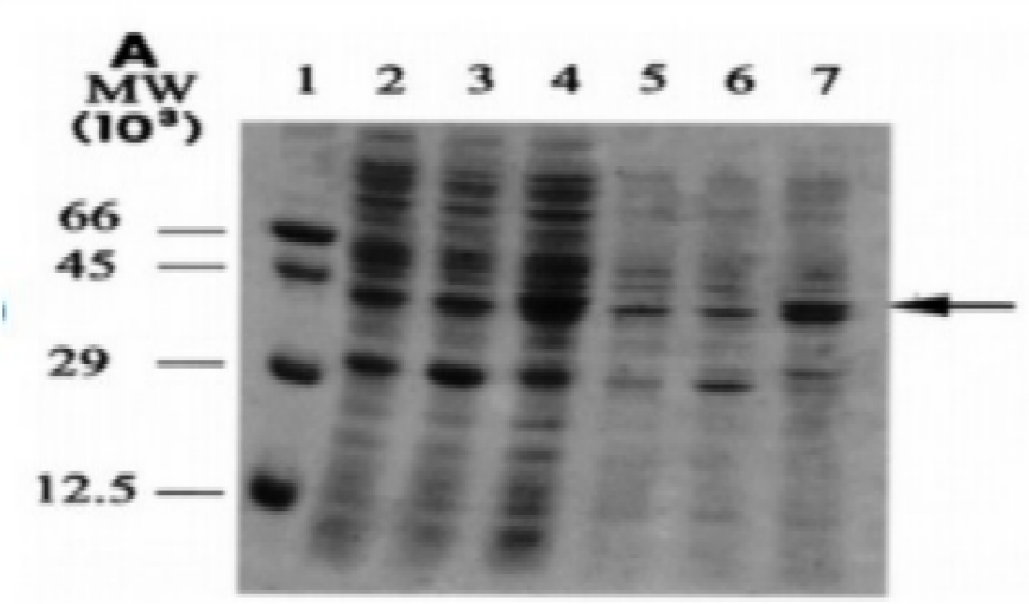
The data in Figures 1-4 comprehensively depict the experimental results of the molecular cloning and protein expression investigations. The results show that recombinant DNA technology could amplify, clone, and express the GST protein. Figure 1 depicts the PCR amplification of GST DNA as a single band with an expected size of 480 bp at lanes 2,3,4,5,6,7,8,9, and 15. The target DNA was amplified effectively, demonstrated by the strong and distinct band intensity at lanes 2,3,4,5,6,7,8,9, and 15 (Wang et al., 2020). This discovery is crucial because strong PCR amplification guarantees enough target DNA is available for subsequent molecular cloning and expression investigations. The colony PCR in Figure 2 shows the expected DNA band sizes (approximately 200 to 600 bp) in lanes 1 to 5, confirming successful cloning of the GST DNA into the plasmid vector. This proves that the bacterial cells successfully created and multiplied the recombinant plasmid encoding the GST gene. Figure 3 illustrates the mapping of plasmid DNA by restriction enzymes, demonstrating that the GST DNA insert is present in the plasmid vector. The GST DNA was successfully inserted into the plasmid vector since the expected fragment sizes were seen following digestion with the selected restriction enzymes as seen at lane 2,3,4,5, and 6 which showed sizes of between 480bp to 16210 bp (Green & Sambrook, 2021). The restriction endonucleases mapping is crucial in ensuring that recombinant plasmids are built correctly and contain the intended insert. The appearance of a prominent band with a characteristic molecular weight of around 35–40 kDa shows that the produced GST protein has been effectively purified and is present in sizable proportions. The produced protein’s identification is further supported by the expected molecular weight of the GST protein, which is in the region of 35–40 kDa and matches the size of the observed band. Notably, the lack of a band at 26 kDa implies that the purification procedure successfully removed any impurities or unattached protein. This outcome demonstrates how precise and effective the purifying technique was. The data allowed for the precise production of extracts from the bands with the highest relative masses. Both hosttransformed plasmids and non-transformed cells were absent. The percentage of bacterial culture solubility in the recombinant protein is estimated using SDS-PAGE; a great deal of fusion was thus represented. As a result, the findings in Figures 1-4 show that the GST DNA was successfully molecularly cloned and translated into a protein using recombinant DNA technology (Sakhel et al., 2021). The GST protein’s effective amplification, cloning, and expression lay the groundwork for future molecular biology studies and experiments.
References
Green, M.R. and Sambrook, J. (2021) “A guide to cloning the products of polymerase chain reactions,” Cold Spring Harbor Protocols, 2021(9). Web.
Sakhel, B. et al. (2021) “Simplification of the purification of heat stable recombinant low molecular weight proteins and peptides from GST-fusion products,” Journal of Chromatography B, 1172, p. 122627. Web.
Simmet, K., Wolf, E. and Zakhartchenko, V. (2020) “Manipulating the epigenome in nuclear transfer cloning: Where, when and how,” International Journal of Molecular Sciences, 22(1), p. 236. Web.
Wang, X. et al. (2020) “A highly integrated system with rapid DNA extraction, recombinase polymerase amplification, and lateral flow biosensor for on-site detection of genetically modified crops,” Analytica Chimica Acta, 1109, pp. 158–168. Web.