Abstract
The project Lab Experiments 6-10: Yeast DNA Cloning and Analysis is a 5-session lab and bioinformatics exercise created to familiarize learners with ideas and procedures in recombinant DNA technology, genomics, and bioinformatics. The project focuses on cloning and sequencing genomic DNA from Saccharomyces cerevisiae, the budding yeast, using techniques such as ligation, analytical endonuclease enzyme digestion, plasmid DNA minipreps, E. coli transformation, DNA sequencing, agarose gel electrophoresis, BLAST, and database searching and interpretation.
The ultimate objective is to identify and characterize the genomic DNA of S. cerevisiae, ascertain whether it encodes proteins, and determine whether related or equivalent genes exist in other species. The laboratory is divided into sections. The first section involves digesting genomic DNA from the budding yeast Saccharomyces cerevisiae with the EcoRI restriction enzyme and purifying the DNA using a Qiagen column purification method.
In the second step, the purified DNA must be ligated into a plasmid vector to be transformed into competent E. coli cells. Plasmid DNA miniprep, followed by electrophoresis of EcoRI-cut and uncut plasmid samples, is the sequencing inserts and bioinformatics step in the lab’s final process. The lab aims to provide students with practical experience in molecular biology techniques and help them gain a deeper understanding of how DNA manipulation works.
Introduction
The five sessions for this experiment in the lab aim to decipher the Saccharomyces cerevisiae genome, a type of yeast that buds. Sessions 6 through 10 of the Lab Experiments are the ones that apply to this project (Laboratory Manual). A group of four students will be assigned to each lab bench and complete the assigned work. Like mammals, yeasts are eukaryotic organisms with similar genetic structures and regulatory systems.
However, yeasts can be used to study fundamental biological processes shared by all eukaryotes because they have a more straightforward genome and are simpler to work with in a lab. By studying these processes in yeast, researchers can gain insights into how they may work in more complex organisms, such as mammals. Thus, the eukaryotic organism’s yeast (S. cerevisiae) is, most likely, the tool that can be evaluated as the simplest for researching mammalian gene regulation.
The DNA molecule’s structure consists of repeating units called nucleotides, is relatively uniform, and lacks the distinguishing features that make other biological molecules, such as proteins or RNA, more amenable to analysis. Hence, it was the most challenging genetic factor to understand in the early 1970s. Several technological developments in the mid-1970s made it possible for scientists to research and work with DNA more conveniently.
For instance, the discovery of endonucleases made it possible to cut DNA into exact fragments. The advancement of DNA sequencing methods allowed scientists to pinpoint the exact order of the nucleotides in a DNA molecule. The contemporary phase of genetic research was made possible by these developments, along with the invention of recombinant DNA technology, which allowed scientists to combine DNA from various sources and produce novel genetic constructs.
DNA has become one of the most thoroughly studied molecules in biology, and techniques for manipulating and analyzing DNA are now widely used in genetic research. Only the analysis of proteins or RNA sequencing was capable of studying it. To analyze and change it, however, is currently the most straightforward approach.
Genomic DNA can be isolated by digesting yeast cells with an enzyme or by breaking up the cells with tiny glass beads. Cells are continuously frozen and thawed in lysis buffer to break down the cell wall and release the genomic DNA. Genomic DNA is extracted, isolated, and precipitated using ethanol following cell lysis. Restriction enzymes, such as Alu1, EcoRI, and BamHI, can digest genomic DNA. Each digest is loaded with dye after digestion, and the digests are then incubated and cooled.
Genomic DNA that has been digested is loaded onto an agarose gel and subjected to electrophoresis; the results are then analyzed. Sanger invented the most straightforward sequencing technique, and other techniques have since been developed, including next-generation sequencing (NGS) methods that utilize technologies from companies such as Illumina, PacBio, and Oxford Nanopore. It is now possible to isolate specific regions of the DNA genome and replicate them billions of times.
A plasmid is an extrachromosomal double-stranded DNA that can reproduce itself in bacteria. They are known as vectors because they are essential elements in recombinant DNA technology that introduce various DNA segments to target organisms. By increasing the number of recognition sites, they have been further modified to enhance their utility and can now simultaneously carry multiple DNA strands. Therefore, this report describes the creation and analysis of recombinants.
Experimental Equipment
- Micropipettes (0-20 µL, 20-200 µL, and 100-1000 µL).
- Sterile tips for pipetting.
- Markers for labeling.
- Vortexes.
- Microcentrifuge.
- Water bath.
- 37°C incubator with shaker.
- Agarose gel electrophoresis box and power supply.
- -80°C freezer (for storing competent cells).
- Glass spreaders (for plating cells).
- Bunsen burners, ethanol (for sterilizing glass spreaders).
- EcoRI restriction enzyme.
- Yeast genomic DNA.
- Plasmid vector.
- Competent E.coli cells.
- LB agar plates containing antibiotics (for bacterial selection).
- Qiaquick PCR purification kit.
- QIAprep Miniprep kit.
Procedure
The EcoRI restriction enzyme was used to break down a sample of yeast genomic DNA. Each group received 10 μL of yeast genomic DNA at a concentration of 275 ng/μL, 1.5 μL of deionized water, 2.5 μL of 10x EcoRI buffer, and 11.5 μL of EcoRI (10 units/μL). After that, the mixture was incubated for 30 minutes at 37°C. After incubation, the broken-down DNA was purified using the QIAquick purification procedure, and the DNA fragments were visualized by gel electrophoresis.
Following digestion, the digested DNA was loaded onto an agarose gel and subjected to electrophoresis. DNA was purified using the Qiagen protocol while the gel was running. The QIAquick PCR Purification Kit was used to purify the digested DNA, and the purified DNA was then observed using a spectrophotometer. 20 μL of a digestion reaction mix containing 2.2 pg of EcoRI-cut DNA for each group was the initial starting material. 30 μL of elution was used after the mixture was diluted and loaded onto a Qiagen column. The tube was kept at -200°C, and the anticipated concentration of the eluate was 22–44 ng/l.
The purified DNA and a plasmid vector were used for a ligation reaction. The ligated DNA was then used to transform competent E. coli cells, which were plated onto special agar plates for overnight incubation at 37°C. The digested DNA (the insert) and a vector were combined in a 1:1 ratio during the ligation procedure. T4 DNA ligase and ATP were added to the mixture, which facilitated the formation of covalent bonds between the insert and vector. Competent E. coli cells were subsequently transformed using the ligation mixture, enabling the replication and amplification of the recombinant plasmid DNA.
The QIAprep Miniprep kit was used to perform a plasmid DNA miniprep on the EcoRI-cut and uncut plasmid samples in subsequent sessions. After that, to remove the genomic DNA and cell debris, buffer N3 was added. The plasmid DNA bound to the column matrix after the cleared lysate was transferred to a spin column and centrifuged.
Plasmid DNA was eluted from the column using buffer EB after being washed with buffer PB. The DNA was sent to Psomagen for sequencing, and a spectrophotometer was used to determine the concentration and purity of the plasmid DNA. Following that, the EcoRI restriction enzyme was used to digest the purified plasmid DNA samples, which were then loaded onto an agarose gel and subjected to electrophoresis.
The DNA bands were observed using a UV transilluminator, and the band patterns were compared to the original genomic DNA to evaluate the efficiency of the restriction enzyme digestion. Following EcoRI restriction digestion, the purified DNA is examined by agarose gel electrophoresis. Based on the gel’s findings, insert-containing plasmids digested with EcoRI, electrophoresed wk4, are chosen for industrial sequencing. Then, using the Yeast Genomic DNA database or the yeast sequences in the NCBI GenBank database, BLAST analysis was employed to determine their sequences.
Results
Discussion
Molecular cloning is a method for amplifying and purifying particular DNA sequences. Recombinant DNA technology involves modifying DNA to create recombinant molecules with specific sequences. Molecular cloning involves inserting the desired DNA fragments into a plasmid vector to replicate them in a host cell (Green and Sambrook). Scientists can combine sequences from various sources to produce chimeric genes through recombinant DNA technology.
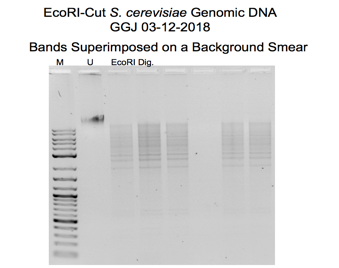
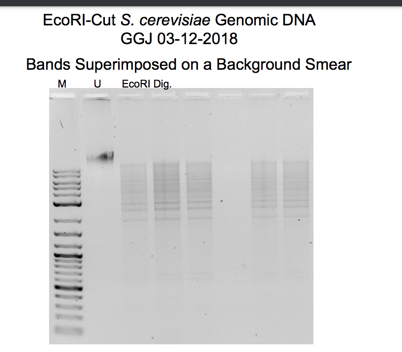
A sample of yeast genomic DNA is first digested with the restriction enzyme EcoRI in the molecular cloning procedure. An analytical gel is used to visualize the size and distribution of the resulting DNA fragments. The digested DNA is run through an analytical gel to confirm the effectiveness of the endonuclease digestion and determine the size of the DNA fragments. The gel displayed bands of various sizes corresponding to the fragments (Lee et al., 2012). The remaining DNA digest can be used later, such as ligation and transformation.
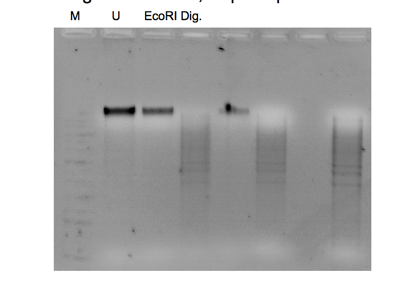
The seen band size on the gel in Figure 1 indicates that some yeast genomic DNA was successfully cut with the EcoRI restriction enzyme. At the same time, some were not based on the outcomes of the gel electrophoresis and DNA purification procedures. The EcoRI-cut DNA was separated and purified from the other reaction components to achieve a final concentration of 40 ng/µL. This step was done using the Qiagen QIAquick DNA purification method. Binding the DNA to a column, washing it to remove impurities, and eluting the purified DNA are all steps in the protocol. The yield of purified DNA was predicted to range from 50 to 100%, so the precise amount of purified DNA in the eluate may differ.
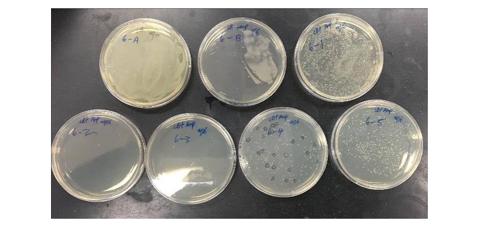
The experiment’s findings demonstrate that E. coli cells can be successfully transformed using a recombinant plasmid that incorporates yeast genomic DNA and vector DNA, as illustrated in Figure 6. The visualization of the DNA fragments using agarose gel electrophoresis and ethidium bromide staining was successful, and the mixture’s marker helped determine the size of the resulting samples.
More colonies were in the plate to which an insert had been added than in the control plate (-), indicating that the insert may have disrupted the function of the plasmid, such as by disrupting a selectable marker, resulting in fewer colonies in the control plate. The positive and negative signs on the Agar plates were used to test the E. coli concentration in the DNA (Lee et al.). Positive indicated an increase in the colonies, which was expected given the growth, while negative meant no increase in the colonies.
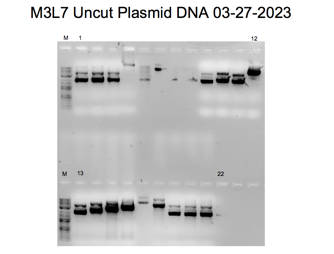
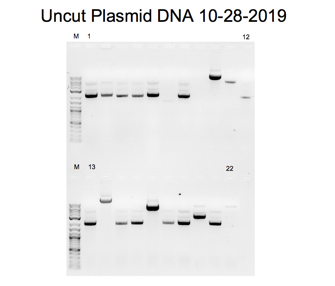
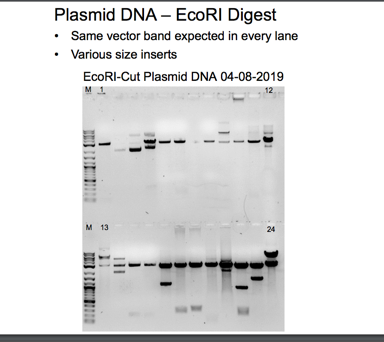
When DNA is bound to ethidium bromide, white bands can be seen when exposed to UV light due to the ethidium’s intercalation into the base pairs. The dominant band in the uncut plasmid lane displayed higher mobility due to the supercoiled nature of the uncut pair, which migrates more quickly than the linear DNA. The electrophoresis diagrams show lanes 7, 10, and 14 appear devoid of inserts (Green and Sambrook). For other band sizes, the marker serves as the reference baseline. The analysis using gel electrophoresis was successful. When the plasmid genetic material was isolated, the uncut sample exhibited a single band with an approximate size of four kilobases.
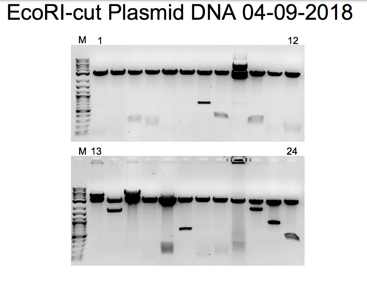
Figure 4 shows the first two bands of the EcoRI-cut Miniprep Gel 1 with digested plasmid and DNA inserts. In the second part of the gel, as shown in Figure 5, the Sample numbers range from 1 to 7. The EcoRI cut Miniprep has samples in the second part of the gel, ranging from 7 to 18. According to the agar plates, the yeast DNA, combined with EcoRI, recreates the original plasmid, and the plasmid miniprep produces new bands.
Due to the presence of DNA from two different organisms—a type from E. coli and another type from yeast—the isolated plasmid is of the recombinant type. Since markers contain DNA fragments of known lengths, they are useful for determining the size of the final samples. The source and vector genetic material samples were combined in a buffer solution containing ATP and DNA ligase.
As a result, a group of recombinant plasmids containing the vector genome and a small piece of yeast genomic DNA was created. The ligation products were combined with E. coli cells that had previously undergone a CaCl2 pretreatment to increase the permeability of their cell membranes and facilitate the uptake of genetic material molecules from the surrounding media. As a result, transformation refers to the process by which DNA enters cells.
The products enabled the bacteria to take up plasmid DNA and replicate, while eradicating those that could not do so. The bacteria were spread on selective LB plates containing ampicillin. Therefore, the resulting colonies were independent and viable. It contains the plasmid DNA that had been successfully transformed into E. coli bacteria that will be incorporated into yeast. The insert is a DNA component backbone in the ligation process.
The two colonies on the (-) plate could have come from various sources, including the following: The amp concentration in the agar plate was not high enough to prevent the colonies from growing. Some of the vector’s phosphatase treatment was probably ineffective, as some could ligate without an insert. This background demonstrates that two or three colonies on the (+) plate have vectors present but without the insert.
Agarose gel electrophoresis is a potent separation technique frequently used when analyzing DNA fragments produced by restriction enzymes. It is a practical analytical technique for breaking down DNA fragments of different sizes, ranging from 100 bp to 25 kb. While pulse-field gel electrophoresis separates DNA fragments larger than 25 kb, polyacrylamide gel electrophoresis is more efficient for separating DNA parts smaller than 100 bp.
Bands on an agarose gel that correspond to the purified plasmid DNA can be seen in Figures 2, 3, 4, 5, 7, and 8. A band of 4kb, consistent with the linearized plasmid DNA, was visible in the EcoRI-cut plasmid samples. In contrast, the circular plasmid DNA should have been visible in the uncut plasmid samples. The plasmid length used in the experiment affected the size of the bands. In molecular biology, plasmids typically range in size from a few kilobases to several tens of kilobases. The exact size of the bands can be estimated by comparing the bands to a DNA ladder run on the same gel with known sizes.
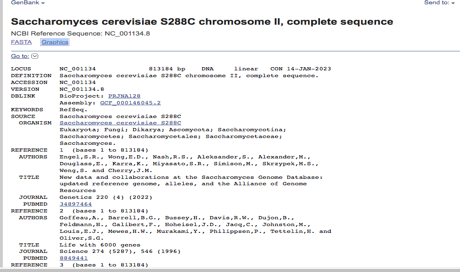
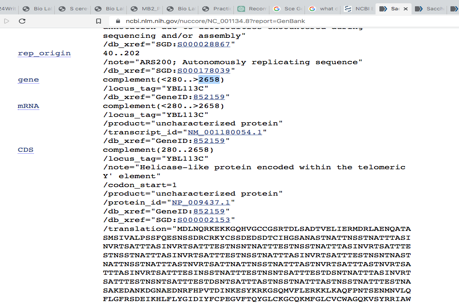
The results of the BlastN algorithm revealed a match between the Cloning vector pGRB2.1 sequence Chromosome II, Complete sequence with coordinates 280>2865, and the query sequence NC_001134: S. cerevisiae S288C chromosome, Chromosome II, as seen in Figures 9 and 10. The ORF sequence for this alignment is transcribed in the Watson (rightward) strand and has the gene name -YBL113C.
Further investigation revealed that the carbohydrate-binding domain (CBD), the gene product of -YBL113C, is in the cell nucleus. The gene product’s function suggests that it may be involved in carbohydrate metabolism. These results offer important new information about the potential function of -YBL113C in Saccharomyces cerevisiae S288C (National Center for Biotechnology Information).
They may help guide future research on the interaction between CBDs and carbohydrate metabolism in yeast cells. The gene at this location, YBL113C, encodes a protein with a predicted molecular weight of 39.6 kDa and is transcribed in the direction of the rightward strand. The protein encoded by YBL113C belongs to the family of CBDs, which are known to contribute to the degradation of plant cell walls and the maintenance of cell wall integrity.
Despite being a gene found in the yeast S. cerevisiae, YBL113C has no known human homolog. Additionally, cellulose-binding domains, or CBDs, are not typically found in humans or other animals, despite their known role in maintaining plant cell wall integrity and degradation. They are instead present in a wide range of microorganisms and fungi.
YBL113C has been discovered to be involved in several additional biological processes, including meiosis and sporulation, in addition to its predicted CBD function. It also interacts with other proteins that regulate transcription, process RNA, and synthesize proteins (National Center for Biotechnology Information). The YBL113C protein product is primarily found in the cell’s nucleus, where it probably carries out its activities related to gene expression.
Conclusion
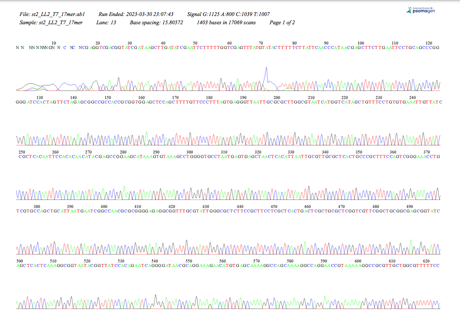
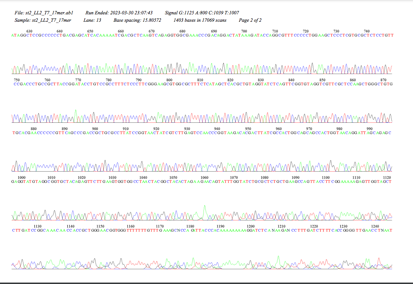
The experiment aids the comprehension and in-depth analysis of the ligation method and gel electrophoresis. Cloning yeast genomic DNA, identifying genes from a fragment, and determining a gene’s human homology are all steps that must be taken. The alignment with the chromosomal sequence was achieved by analyzing the EcoRI-cut S. cerevisiae Genomic DNA (M3L7, 03-13-2023). Re-digested yeast DNA was ligated into a plasmid vector, which gave E. coli the ability to resist ampicillin.
Plasmid DNAs from those that passed the E. coli Transformation, Minipreps & RE Screening were chosen and sent for sequencing. The BLAST search process utilized plasmids from the yeast results. The sequence found at coordinates 280–2865 on Chromosome II aligned with the gene YBL113C, which encodes a CBD protein located in the nucleus, according to the Saccharomyces cerevisiae S288C chromosome’s BlastN analysis. According to the analysis, the sequence is an open reading frame and is transcribed in the Watson strand direction. The CBD protein facilitates beta-glucan biosynthesis, a crucial element of the yeast cell wall.
Works Cited
Bio 301, Laboratory Manual. (2023). Yeast DNA Cloning and Analysis.
Green, Michael R., and Joseph Sambrook. “Analysis of DNA by Agarose Gel Electrophoresis.” Cold Spring Harbor Protocols, vol. 2019, no. 1. Web.
Lee, Pei Yun, et al. “Agarose Gel Electrophoresis for the Separation of DNA Fragments.” Journal of Visualized Experiments, no. 62, 2012. Web.
National Center for Biotechnology Information. “Saccharomyces Cerevisiae S288C Chromosome II, Complete Sequence – Nucleotide – NCBI.” National Center for Biotechnology Information, U.S. National Library of Medicine. Web.