Introduction
This lab report aims to give a general overview of the Baculovirus Expression Vector System (BEVS) and how it expresses a eukaryotic 3′ tRNA processing enzyme in a recombinant fashion. It describes the lytic lifecycle of baculoviruses, their structure, and how highly concentrated recombinant proteins can be made using them.
Theory
The Baculoviridae family of enveloped double-stranded DNA viruses, or baculoviruses, is a diverse group that is only pathogenic to arthropods, primarily lepidopteran insects (moths and butterflies). Taxonomically, they are separated into two genera: granulovirus (GV) and nucleopolyhedrovirus (NPV). The diameter and length of virions are approximately 40–50 nm and 200–400 nm, respectively, and their circular DNA genomes can be 80–200 kilobase pairs (kbp) long (Harish et al., 2021). The nucleocapsid is a structure comprising the genome compressed into a “core” within the cylindrical capsids, giving them the name. The structurally different ends of the cylindrical capsids give the virion polarity.
Nucleopolyhedroviruses (NPVs) are lytic viruses grouped inside polyhedra, which are protein structures that resemble polyhedral crystals and are environmentally stable. For instance, dying insects leave these behind on the plants they were feeding on or in the soil (Coulibaly, 2019). The polyhedra will be consumed by the subsequent insect larva that tries to feed there, dissolving in the insect’s slightly alkaline midgut and releasing the infectious virus to start a new round of infection (Coulibaly, 2019). As the infection cycle develops in the cells lining the gut, spreading the infection throughout the insect’s body, new viral particles bud from the plasma membranes of the infected cells.
The polyhedrin protein, which creates the polyhedral structures, is expressed at extremely high levels later in the infection cycle and encloses or “occludes” the virions assembled in the host cell’s nucleus. This creates a fresh batch of polyhedrin-occluded virus particles that can survive in the environment and wait patiently for the subsequent unwary insect larva to consume them.
Yu et al. (2021) showed how the Autographa californica multiple nuclear polyhedrosis virus (ACMNPV) could be used to express large amounts of recombinant proteins. They inserted protein-coding genes into the polyhedrin locus under the transcriptional regulation of the powerful polyhedrin promoter. Studies have shown that it is possible to achieve superior expression of foreign proteins in cell culture by infecting cells at a lower MOI (1) and allowing the virus to spread horizontally for up to 24 hours. The culture is harvested 4-5 days after infection to obtain the desired expressed protein at a high level. The baculovirus expression vector system (BEVS) is an effective tool for eukaryotic cells.
Overall, the system for expressing recombinant proteins using baculovirus expression vectors is dependable and effective. The BEVS enables protein expression in a eukaryotic setting that closely mimics the cellular setting where proteins are usually expressed. We successfully expressed a eukaryotic 3′ tRNA processing enzyme using the BEVS, and our findings agree with earlier research. Thus, researchers investigating various facets of protein expression and function can benefit from using the baculovirus expression vector system.
Materials and Reagent
- Lysis Buffer 100ml
- Wash Buffer 100ml
- Elution Buffer 100ml
- Sodium Dodecyl Sulfate (SDS)
- Falcon-capped centrifuge tubes
- SF9 cells
- Centrifuge
- Protein Loading Buffer (PLB)
- Bacterium
- Cleared Lysate (CL)
- Flow-through (FT)
- SDS-PAGE gels
- Bromophenol blue tracking dye
- Typhoon Scanner
Methodology
The following methods were employed:
- The experiment used SF9 cells, Falcon-capped centrifuge tubes, Protein Loading Buffer (PLB), and bacteria.
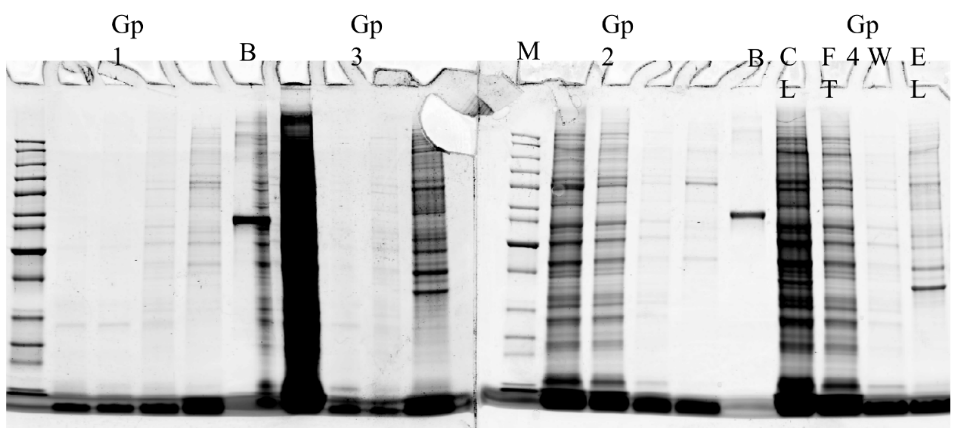
- Protein purification and analysis were performed using Cleared Lysate, Flow-through, SDS-PAGE gels, and Bromophenol blue.
- The target gene was introduced into the cell’s nucleus. Reverse transcriptase converted the DNA into cDNA, which was then used to translate mRNA into proteins.
- Affinity purification was used to purify the proteins.
- The Invitrogen BEVS Manual diagram and the plastic HTa Multiple Cloning Site (MCS) were used to clone and express the target gene.
- Cells were lysed, and the lysate was cleared to remove cellular waste and other impurities.
- The target protein was coupled to a 6XHis-tagged column to identify it as its affinity ligand. An imidazole-containing buffer was used to remove the target protein from the column.
- The expressed protein was detected using fluorescence microscopy and an antibody to the eukaryotic 3′ tRNA processing enzyme.
Procedure and Results
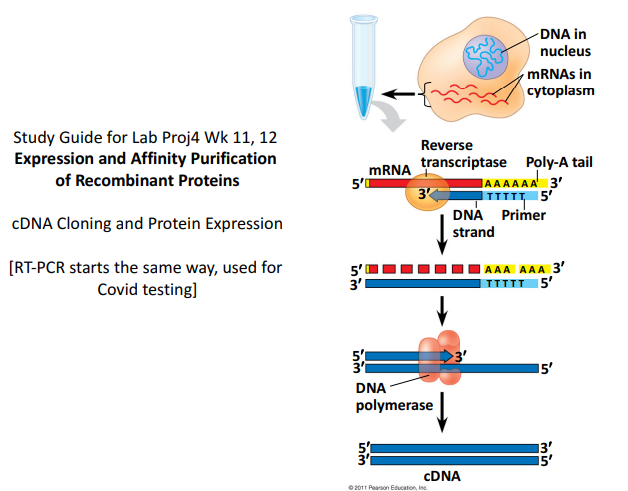
This image depicts the process of producing and purifying recombinant proteins from a eukaryotic cell. The procedure begins with introducing the target gene into the cell’s nucleus. Subsequently, the reverse transcriptase enzyme is used to convert this DNA into cDNA. Proteins are produced following the translation of the cDNA into mRNA. Finally, affinity purification methods can be used to clean these proteins. Any desired protein can be expressed and purified using this method.
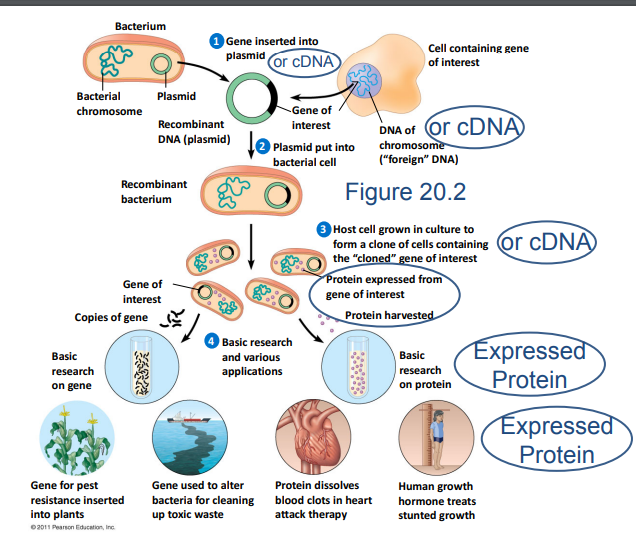
This figure illustrates the process of recombinant protein expression using the Baculovirus Expression Vector System. A gene of interest is inserted into the polyhedrin locus of the baculovirus genome under the transcriptional regulation of the strong polyhedrin promoter. The recombinant virus is then used to infect insect cells, such as Sf9 cells, which express the gene of interest. The expressed protein is harvested from the infected cells at 72 hours post-infection.
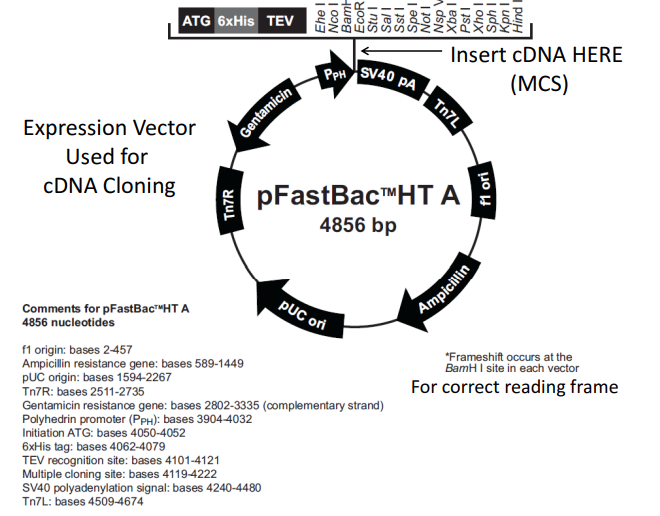
The expression vector used for cDNA cloning is shown in this figure. A multi-cloning site, an enhancer/promoter, a selectable marker, and an origin of replication are all components of the vector that are required for a successful cloning procedure. A DNA sequence known as the multi-cloning site makes it simple to introduce a foreign gene or cDNA. A DNA sequence known as an enhancer or promoter encourages the expression of a gene of interest. An antibiotic-resistant gene serves as the selectable marker, enabling the identification of cells that have taken up the vector (Dormatey et al., 2021). A DNA sequence that enables the replication of the vector inside the host cell is the replication source.
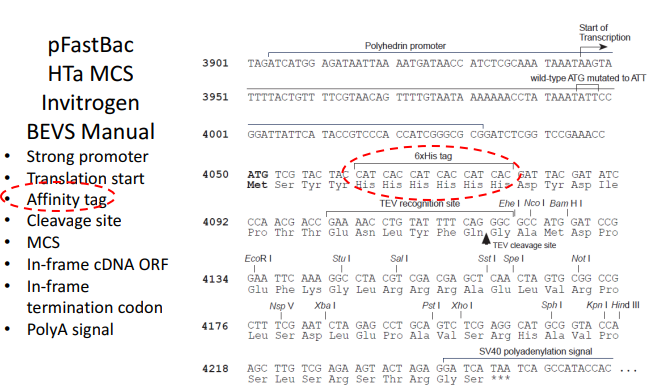
The plastic HTa Multiple Cloning Site (MCS) is shown schematically in the figure from the Invitrogen BEVS Manual. It contains the potent polyhedrin promoter to drive the desired protein’s expression. Along with the affinity tag, which is used to purify the expressed protein, it also contains the translation start site, the first codon of the protein-coding region. It also contains the cleavage site, where the expressed protein’s affinity tag is cut off, and the MCS, a collection of particular restriction enzyme sites that can be used to splice in foreign DNA. Finally, it contains the polyA signal, indicating the completion of transcription, the in-frame cDNA open reading frame (ORF), the in-frame termination codon, and the beginning and end of the protein-coding sequence.
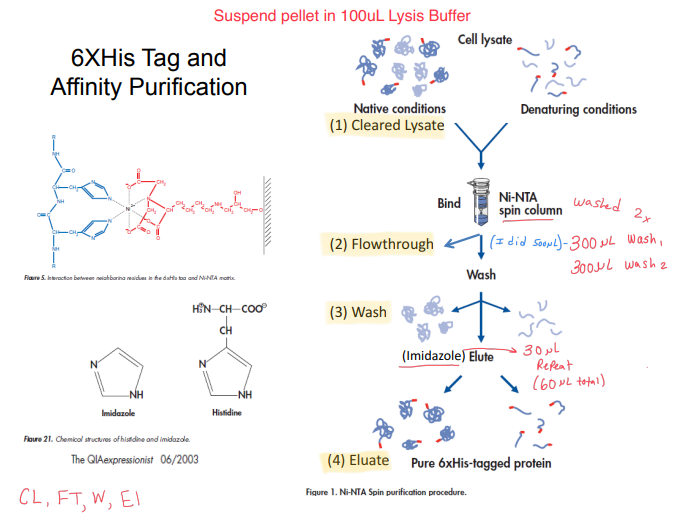
- The cell pellet was suspended in 100 µl of lysis buffer to dissolve the cell walls and make the proteins inside accessible. This step entails suspending the cell pellet in the lysis buffer. Detergents and protease inhibitors found in the lysis buffer aid in solubilizing the proteins and preserving their integrity.
- 6XHis Tag and Affinity Purification: The target protein is bound to a column treated with a 6XHis tag to identify it as the target protein’s affinity ligand. After that, the target protein is removed from the column using an imidazole-containing buffer.
- Cleared Lysate: The lysate, a liquid sample containing the extracted proteins, is cleared in this step. This step is essential to eliminate any cellular waste or other contaminants that might obstruct the purification of the protein.
- Flowthrough: The flowthrough from the affinity purification column and 6XHis tag is collected at this stage. The proteins that did not bind to the 6XHis tag affinity ligand are in the flowthrough and can be discarded or treated further.
- Wash: In this step, an imidazole-containing buffer is used to wash the column. This step is required to eliminate any proteins that are bound inadvertently and to improve the specificity of the 6XHis tag affinity purification.
- Elute: In this step, an imidazole-containing buffer is used to elute the target protein from the column. To extract the desired protein from the column, this step is required.
This figure shows the immunofluorescence analysis of the expressed protein. The expressed protein is detected using an antibody specific to the eukaryotic 3′ tRNA processing enzyme and visualized using fluorescence microscopy.
Conclusion
Baculoviruses are double-stranded DNA viruses that are enveloped and only pathogenic to arthropods, to sum up. A valuable tool for expressing recombinant proteins in eukaryotic cells is the Baculovirus Expression Vector System (BEVS). BEVS can express an eukaryotic 3′ tRNA processing enzyme that can be harvested 72 hours after infection. Additionally, protein expression can be improved by infecting cells at a lower multiplicity of infection and allowing the budding virus to spread throughout the culture. This lab report has shown that proteins can be expressed using the baculovirus expression vector system in a cellular-like eukaryotic environment.
References
Coulibaly, F. (2019). Chapter Nine – Polyhedra, spindles, phage nucleus, and pyramids: Structural biology of viral superstructures (F. A. Rey, Ed.). ScienceDirect; Academic Press. Web.
Dormatey, R., Sun, C., Ali, K., Fiaz, S., Xu, D., Calderón-Urrea, A., Bi, Z., Zhang, J., & Bai, J. (2021). ptxD/Phi as alternative selectable marker system for genetic transformation for bio-safety concerns: A review. PeerJ, 9, e11809. Web.
Harish, S., Murugan, M., Kannan, M., Parthasarathy, S., Prabhukarthikeyan, S. R., & Elango, K. (2021). Entomopathogenic viruses. Microbial Approaches for Insect Pest Management, 1–57. Web.
Yu, Q., Chang, P., Liu, X., Lü, P., Tang, Q., Guo, Z., Qiu, J., Chen, K., & Yao, Q. (2021). Bombyx mori Pupae efficiently produce recombinant AAV2/HBoV1 Vectors with a Bombyx mori Nuclear Polyhedrosis Virus Expression System. Viruses, 13(4), 704. Web.