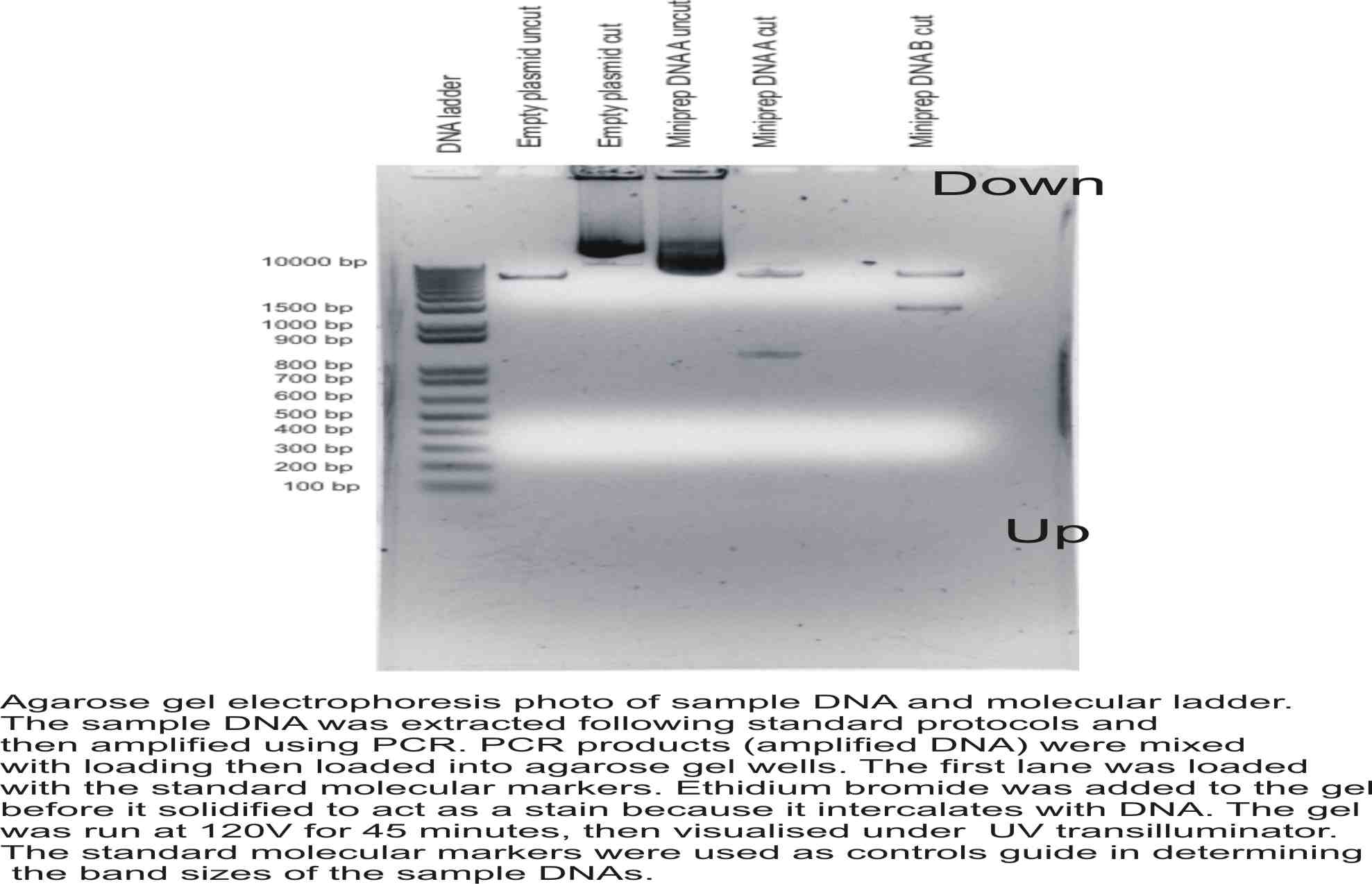
Produce a figure with an appropriate figure legend of your agarose gel that could be published in a scientific journal
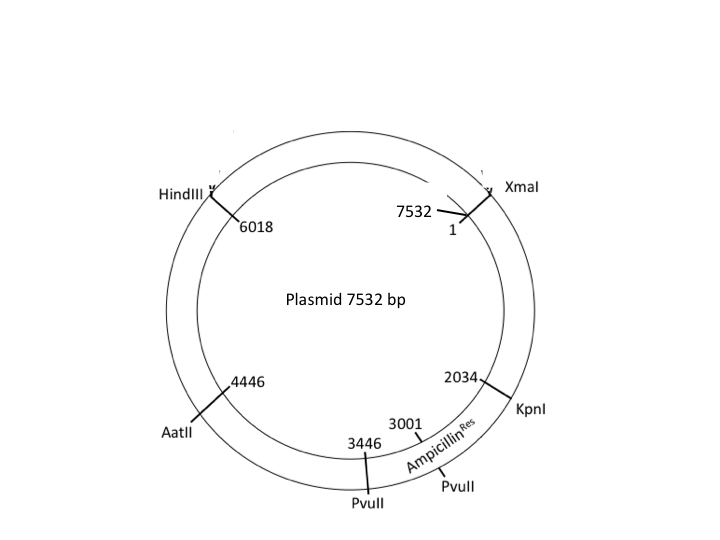
A map of a plasmid
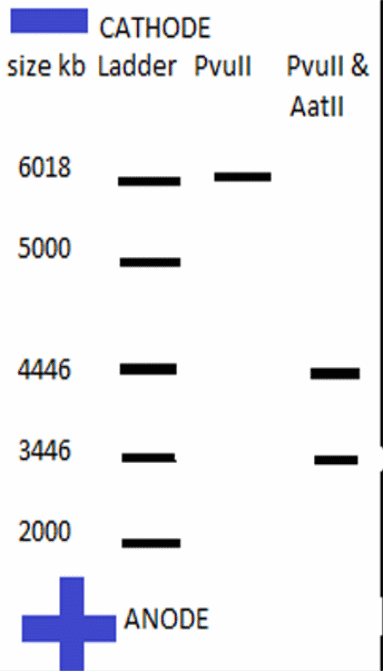
- Produce a ‘virtual’ agarose gel(i.e. use PowerPoint to draw a gel picture that resembles the ‘real’ gel that you show in question 1 above). The virtual gel should show the band pattern that would result from incubating the plasmid with restriction enzymes as indicated below (assume complete digestion of all available sites). Simply draw black lines to represent the resulting DNA fragments. Indicate the ‘+’ and ‘-‘ pole of the gel apparatus/DNA migration.
- The virtual gel should show the following three lanes:
- lane 1: show migration of a suitable DNA ladder (simply ‘invent’ marker bands of informative size and indicated their size on the virtual gel).
- lane 2: show band patterns resulting from digest with the HindIIIendonuclease
- lane 3: show band patterns resulting from digest with both PvuII andAatIIendonculeases
- Commercially available restriction endonucleases like the ones that are used in the prac are typically obtained from bacteria. The bacteria use those enzymes to destroy foreign DNA in case they are attacked e.g. by ‘invading’ viruses. Why do the enzymes not destroy the own DNA of the bacteria? (1 mark)
- Bacteria have their protection mechanisms that protect them from being degraded by endonucleases. This is achieved by the addition of methyl groups on a few bases on each strand. The methylation process is carefully done to avoid interfering with the normal base pairing of nucleotides. Once the bases are methylated, the restriction enzymes produced by the bacteria will not attack its DNA. However, any foreign DNA molecule that gains access to the bacterium will be destroyed by the digestive enzymes (endonucleases). The protective mechanism by bacteria has made it possible for applications of restriction enzymes on bacterial plasmids without the fear of interfering with plasmids’ DNA. Therefore, plasmids are successfully cleaved to produce sticky ends for insertion of the gene of interest.
The invention of ‘recombinant insulin’ facilitated the economic supply of large amounts of the hormone to patients suffering from diabetes
- Why is it possible that the gene expression machinery of bacteria can use the human insulin mRNA to correctly synthesise human insulin polypeptides?
- The recombinant DNA technology uses insulin-coding genes which are inserted into self replicating plasmids. In order for the insulin-coding gene to fit into the plasmid, both the plasmid and the gene are digested or cut with the same restriction enzyme that leaves sticky ends on their edges. Some of the restriction enzymes are HindIII and ECOR1. The sticky ends enable the plasmid and the foreign insulin DNA molecule to base pair at their open ends, forming stable covalent bonds. The plasmid vectors, now with the insulin gene inserts, are introduced into E. coli bacteria where they self-replicate. Also present in the growth media are the selectable markers which show the success or failure of the cloning process. During the replication process, the plasmids’ genomic machinery is expressed, and the inserted insulin gene is also expressed. An example of a selectable marker is ampicillin antibiotic resistance gene. The insulin gene expression product is the insulin (protein) which is used to manage patients suffering from diabetes mellitus. The correct insertion of the insulin mRNA into the coding regions of the expression vectors makes it possible to have its gene product expressed. The human insulin polypeptides produced in the insulin recombinant technology process are isolated from reactors and purified for use by diabetic persons.
- Recombinant insulin protein must be correctly folded to be able to interact with insulin receptor such that it can control carbohydrate metabolism in human cells. How is it possible that recombinant insulin is correctly folded when isolated from bacteria?
- Folding of proteins is crucial in their biological functions. The folding makes proteins assume their structural conformations. The four structural conformations of proteins are the primary structures, secondary structures, tertiary structures and the quaternary structures. When polypeptides fold, they form stable three-dimensional structures. What determine the degree of folding and structures formed are the amino acids present in the protein. In addition, research has shown that external factors also determine the type of folding in proteins. Some of these external factors are the temperature of the surrounding and the concentration of molecules in the immediate surrounding. These factors have different effects on the folding of proteins. It has been demonstrated that most of the functional motifs of a protein are found within protein folds. Protein motifs are the functional sites within a protein. For example, the functional unit of an enzyme is its binding site.
Circular plasmid DNA is massively amplified through growth of exponential E. coli cultures. The plasmid preparation that you produced during the prac is, however, largely free of chromosomal E.coli DNA.
- Plasmids were isolated from membranes of the E. coli bacteria by adding high concentration of alkaline solution. The alkaline solution detached plasmid DNA from the E. coli cells leaving chromosomal bacterial DNA attached on the cellular membranes. The plasmid preparation produced in the practical was, therefore, free from chromosomal DNA. Other nucleic acids lysed from the bacteria were ribosomal and transfer RNAs. To destroy most of the RNAs high concentration of alkaline should be used to lyse bacterial cells. RNA destruction occurs due to hydrolysis of the 2’ hydroxyl group in the RNA structure.
In part D of this practical you determined the concentration of nuclei acids in the DNA minipreps using a spectrophotometer in the UV range (260nm).
- Which component(s) of the DNA is/are absorbing in the UV range?
- The nitrogenous bases in DNA molecules absorb UV-light at 260 nm. The nitrogenous bases of DNA enable visualisation of DNA in UV transilluminator following agarose gel electrophoresis. The chemical properties of the nitrogenous bases enable them absorb UV-light
- Considering that DNA absorbs UV-light, what do you think can happen to the DNA of your skin cells when you take a sun-bath at the beach?
- Nitrogenous bases in DNA absorb UV-light at specific wavelength (at 260 nm). Taking a sun-bath at the beach would expose skin cells to the UV-light. The DNA molecules in the skin cells would absorb UV at 260 nm. UV-light introduces changes in the DNA of the skin cells making them have altered gene expression and control patterns. Cells whose genes are expressed constitutively are abnormal cells without growth arresting factors. Therefore, the cells grow and multiply abnormally. Continuous exposure would result in skin cancer because DNA will have mutated to high levels. This has been demonstrated through research in carcinogenesis in persons living in locations with high amounts of UV-light.
- To remove all contaminating RNA from your DNA miniprep you used a buffer containing RNase A. Alternatively, you can destroy RNA by treating your sample with alkaline pH. Why does the latter treatment destroy the RNA but leaves the integrity of the plasmid DNA intact?
- RNAse is an enzyme used to destroy RNA for biological practical purposes. Alkaline pH solution is also used to remove unwanted RNA molecules in biological samples. RNA molecules have 2’ hydroxyl group in their moiety. This is not the case with DNA. The alkaline solution acts on the 2’ hydroxyl group on the RNA molecules thereby destroying the structural and chemical structure of RNA. However, plasmid DNA will remain intact because it does not contain 2’ hydroxyl group in its moiety. Biological scientists have been able to isolate DNA from a mixture of DNA and RNA by treating the mixture with alkaline pH solution. High purity DNA is obtained though such isolation process.