Introduction
An exoenzyme, also known as an extracellular enzyme, is an enzyme that is released into the environment by a cell and acts outside of that cell. Exoenzymes may be generated by both bacterial prokaryotes and eukaryotes. These enzymes are often engaged in the degradation of more significant macromolecules. The breakdown of these macromolecules is necessary for their smaller components to enter the cell via the cell membrane (Ford, 2019). Microbes produce exoenzymes to metabolize substances in their surroundings, and these microorganisms may be used in biochemistry assays to evaluate the presence and activity of exoenzymes. Certain infectious bacteria utilize exoenzymes as virulence agents to help in their proliferative processes. Examples of extracellular enzyme production tests include: Start tests for amylase, Casein hydrolysis tests, Blood gar tests, and DNase tests.
Objectives
- To differentiate organisms based on the production of amylase enzyme
- To differentiate organisms based on the production of protease enzyme
- To differentiate organisms based on the production of hemolysin enzyme
- To differentiate organisms based on the production of deoxyribonuclease enzyme
Materials
- Inoculating wire
- Bunsen burner
- Microscope 4x, 10x, 40x, 100x
- Incubator
- Bacteria: E.coli, Bacillus Subtilis, S.aureus, and S. epidermidis.
- Media: DNase Agar Medium, Blood Agar Medium, Casein (Milk)Test Medium, and Starch Agar.
Methods
Start Agar
Using a sterile technique, a single streak inoculation of Bacillus subtilis on the right and E.coli on the left were done into the center of labeled plate. The plates were then incubated for 24 hours at 37°C. Following incubation, the surface of the plates was flooded with iodine solution via a dropper for 30 seconds. The excess iodine was then poured off and the results for clear colonies around the line of bacterial growth examined.
Casein Test
Using a sterile technique, a single streak inoculation of Bacillus subtilis on one plate and E.coli on another plate were done into the center of labeled plates. The plates were then incubated at 37°C for 24 hours. The milk agar plates were then observed for the presence or absence of clear area or zone of proteolysis, surrounding the growth of each of the bacterial test organisms.
Blood Agar Test
Using a sterile technique, a single streak inoculation of S.aureus on the right and S. epidermidis on the left were done into the center of labeled plate. The plates were then incubated at 37°C for 24 hours. The Blood agar plates were then observed for the presence or absence of a clear (transparent) zone surrounding the colonies, a green, opaque zone, or no notable zones around the colonies.
DNase Test
Using a sterile loop, a single streak inoculation of S.aureus on one plate and S. epidermidis on another plate containing DNase agar were done into the center of labeled plate. The mixture on the plate was then incubated at 37°C for 24 hours. After incubation, the color changes were observed in DNase with methyl green.
Results
Start Agar Test
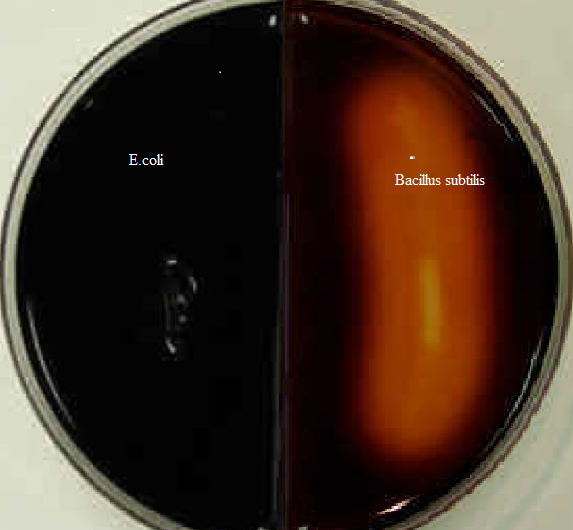
Table 1: Starch test results
Casein Test
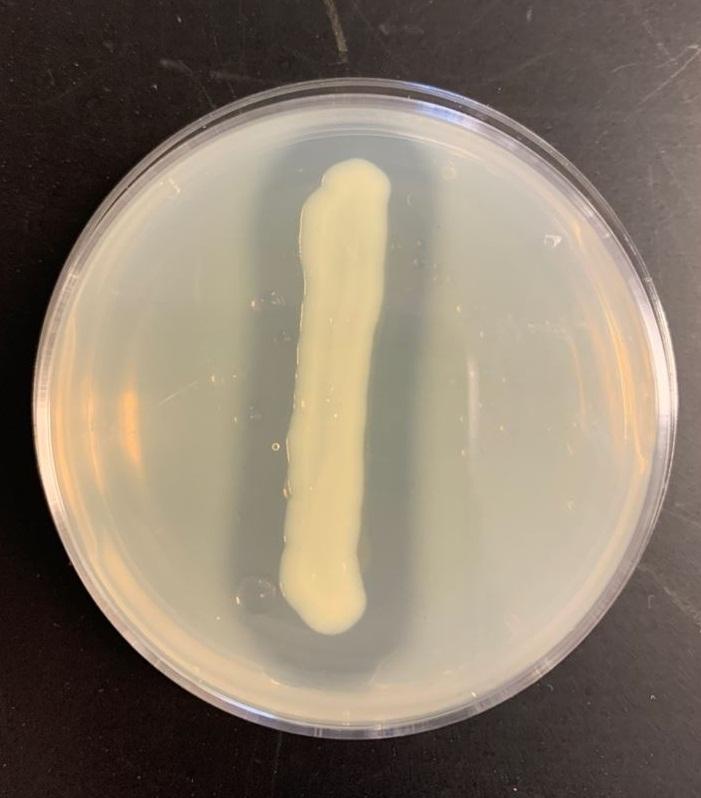
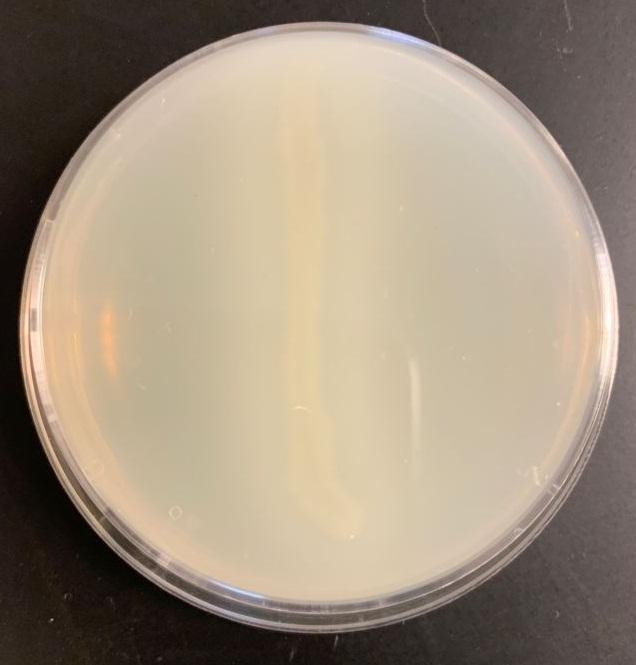
Table 2: Casein test results
Blood Agar Test
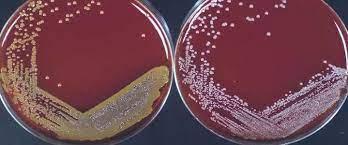
Table 3: Blood agar test
DNase Test


Discussion/Conclusion
Using the enzymes a-amylase and oligo-1,6-glucosidase, the starch test identifies bacteria that can hydrolyze starch (amylose and amylopectin), Often used to distinguish species from Bacillus genera. The enormous size of amylose and amylopectin molecules prevents these organisms from penetrating the bacterial cell wall (Ford, 2019). Bacillus subtilis secretes a-amylase and oligo-1,6-glucosidase into the extracellular space to use these starches as a carbon source. These enzymes break down the starch molecules into smaller subunits of glucose, which may subsequently enter the glycolytic pathway directly. Add iodine to the agar in order to understand the findings of the starch hydrolysis test. E.coli does not release the enzyme a-amylase, iodine combines with starch to produce a dark brown hue. Thus, starch hydrolysis produces a clean zone surrounding bacterial development. Bacillus subtilis has starch hydrolysis activity (see Figure 1, right). E.coli (see Figure 1 on the left) is incapable of hydrolyzing starch.
Bacillus subtilis may break down the casein protein in the Casein test by generating the proteolytic exoenzyme proteinase (caseinase). In order to demonstrate such an action in the laboratory, milk agar is used. The medium consists of nutritional agar supplemented with milk containing casein as a protein substrate (Ford, 2019). Comparable to other proteins, milk protein is a colloidal suspension that imparts color and opacity to the medium by reflecting rather than transmitting light rays. Bacillus subtilis, after inoculation and incubation of agar plate cultures, secretes proteases that display a zone of proteolysis, which is a transparent region around the bacterial growth (see Figure 2). Therefore, transparent halos encircling colonies are suggestive of their capacity to digest casein and result in a clear zone surrounding plated development. The media surrounding the E. coli’s development stays opaque without protease activity (see Figure 3), which is a negative response.
As a differential media, the blood agar test is a rich, complex medium that includes 5% sheep red blood cells. BAP evaluates an organism’s capacity to create hemolysins, enzymes that destroy red blood cells (erythrocytes). S. aureus secretes the enzyme hemolysin, which destroys red blood cells (Ford, 2019). Therefore, S. aureus is a beta-hemolysis because it exhibits complete hemolysis. It is distinguished by a translucent zone encircling the colonies (see Figure 4 left). As a gamma-hemolysis, S. epidermidis does not release any alpha or beta lysine enzyme to degrade RBCs. Thus, white colonies are seen (see Figure 4 right).
Using the enzyme deoxyribonuclease, the DNase test evaluates the hydrolysis of DNA strands. DNases are extracellular endonucleases that cut DNA and liberate nucleotides and phosphate. DNase agar includes nutrients for the bacteria, DNA, and methyl green as the primary indication. Methyl green is a cation that binds to DNA due to its negative charge. S. aureus produces the enzyme deoxyribonuclease to fragment DNA (Ford, 2019). When the DNA is degraded, it can no longer attach to methyl green, and a visible halo will develop in the places where the DNase-producing organism has grown (see Figure 6). However, in S. epidermidis, the enzyme DNase is not generated; thus, there is no DNA degradation; the medium stays green because the methyl green remains bound to the DNA strands (see Figure 7).
Reference
Ford, M. (Ed.). (2019). Medical microbiology (3rd ed.). Oxford University Press.