Introduction
Gene expression can be defined as “the synthesis of structural and functional products from information emanating from genes” (Annis et al. 1939). Most often than not, “the products of gene expression are proteins but in non-protein coding, the product may be a structural RNA” (Annis et al. 1939). The establishment of the exact products of the expression of certain genes calls for specialized molecular analytical procedures. One molecular procedure has been reliable in characterizing genetic make ups of even pathogenic bacteria and viruses to aid in definitive diagnosis; Polymerase chain Reaction (PCR). Advances in technology have developed various forms of PCR meant to address various aspects of gene expression. Such forms include qualitative and quantitative PCR (Life Technologies), which are preferably carried out in real time. This experiment made use of the Real-time quantitative PCR to measure gene expression (Fricker et al. 772).
Real time PCR is one of the most powerful and reliable techniques of gene analysis. It is commonly applied in the quantification of mRNA either from endogenous genes or transfected genes of either stable or transient transfection (Fricker et al. 772). RT-qPCR is arguably the most sensitive technique in the analysis of mRNA yet. When using “real-time PCR to study gene expression, scientists often investigate the decreases and increases in the intensity by which certain genes are expressed” (Livak and Schmittgen 404). This is done by measuring the abundance or deficiency of gene-specific transcripts (Fricker et al. 772). This forms the basis of action for this technique of gene analysis. In this experiment, sample tissues were collected and maintained at favorable conditions to ensure the viability of gens. Genes were extracted from the endoplasmic reticulum, which is located intracellular, of the respective tissue cells.
Endoplasmic reticulum’s (ER) main function is to control protein synthesis and secretion. Additionally, it provides the platform on which secreted proteins are translated, folded and assembled (Bailey and O’Hare 2305). These functions are essential for tissues whose functions involve a lot of secretion activities such pancreatic cells and immunoglobulin (Minematsu, Harumi and Naito 542). It is worth noting that the accumulation of unfolded proteins in the endoplasmic reticulum elicits signal transmission from this organelle to the rest of the cytoplasm and nucleus. The amount of accumulated proteins may exceed signals generated to orchestrate increased chaperone expression. This response involves the transmembrane transcription factor ATF6 and two transmembrane protein kinases; IRE1 and PERK (Shen 761). Folding mechanisms are produced by a number of ERs – resident molecules called chaperones and folding enzymes that form the ER chaperones. The latter is involved in the ER-associated degradation (ERAD) mechanism. “The mammalian transcription CHOP is also known as growth arrest, and the DNA damage gene 153 is encoded by a transcription factor that promotes apoptosis in response to uncontrolled ER stress (GADD153)” (Kroeger et al. 7590). Additional to the ER chaperones and the mammalian transcription CHOP, the ER stress induces the transcription factor x box binding protein (XBP-1), which is also called (TREB5). CHOP and XBP1 are mediated by ERSE. ATF6 is produced as a soluble form found in the nucleus; the soluble ATF6 activates transcription of the CHOP as well as ER chaperone genes and XBP1 (Huang et al. 114)
Gene expression analysis
This can be achieved by a number of methods (Northern Blots, Microarrays, Real-Time PCR), all of which essentially measure the expression of genes by quantifying the mRNA levels for the genes of interest. The method used in this practical is quantitative real-time PCR, high sensitivity, reproducibility PCR-based technique, capable of determining the level of expression of genes from a single cell. Two approaches are often used in analysis of data obtained from gene expression experiments using quantitative real-time PCR technique. They include absolute quantification and relative quantification (Livak and Schmittgen 406). Whereas absolute quantification method provides output by relating the PCR signals obtained to those of a standard curve, relative quantification method does the same by relating the PCR signals obtained to those of the targeted gene transcript (Ballester, Cordón and Folch 1).
Objectives
This experiment had an objective to determine the gene expression levels of the genes encoding CHOP/GADD153, BiP and endogenous control 18S rRNA in treated and untreated cells by perform Real-Time PCR using cDNA sample, to examine the expression.
Results
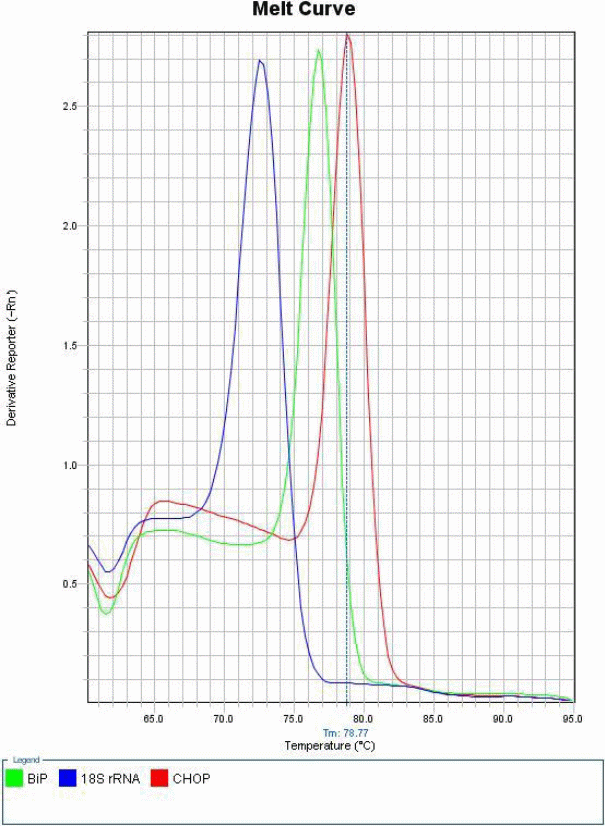
The practical started by making master mixes for CHOP, BiP & 18S rRNA using Sybr Green mix & Gene-specific primers. This was meant to generate a melt curve, which shows the specificity of the experiment method used.
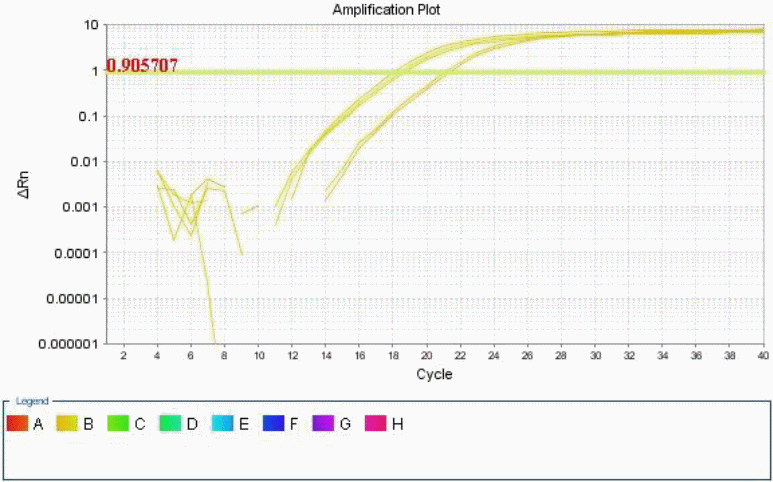
An amplification curve was also generated using the controls. This was meant to establish whether CHOP and BiP are up regulated by Tunicamycin treatment, and what was the level of this up regulation (Ballester, Cordón and Folch 1).
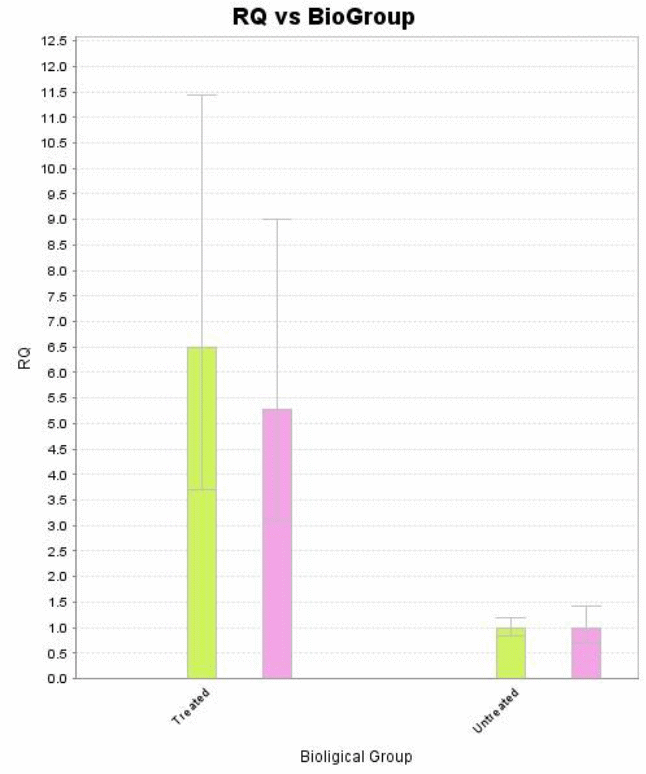
Real-time quantitative PCR was then run using both biological and QR samples of genes, drawing variations between the treated and untreated B104 cells samples.
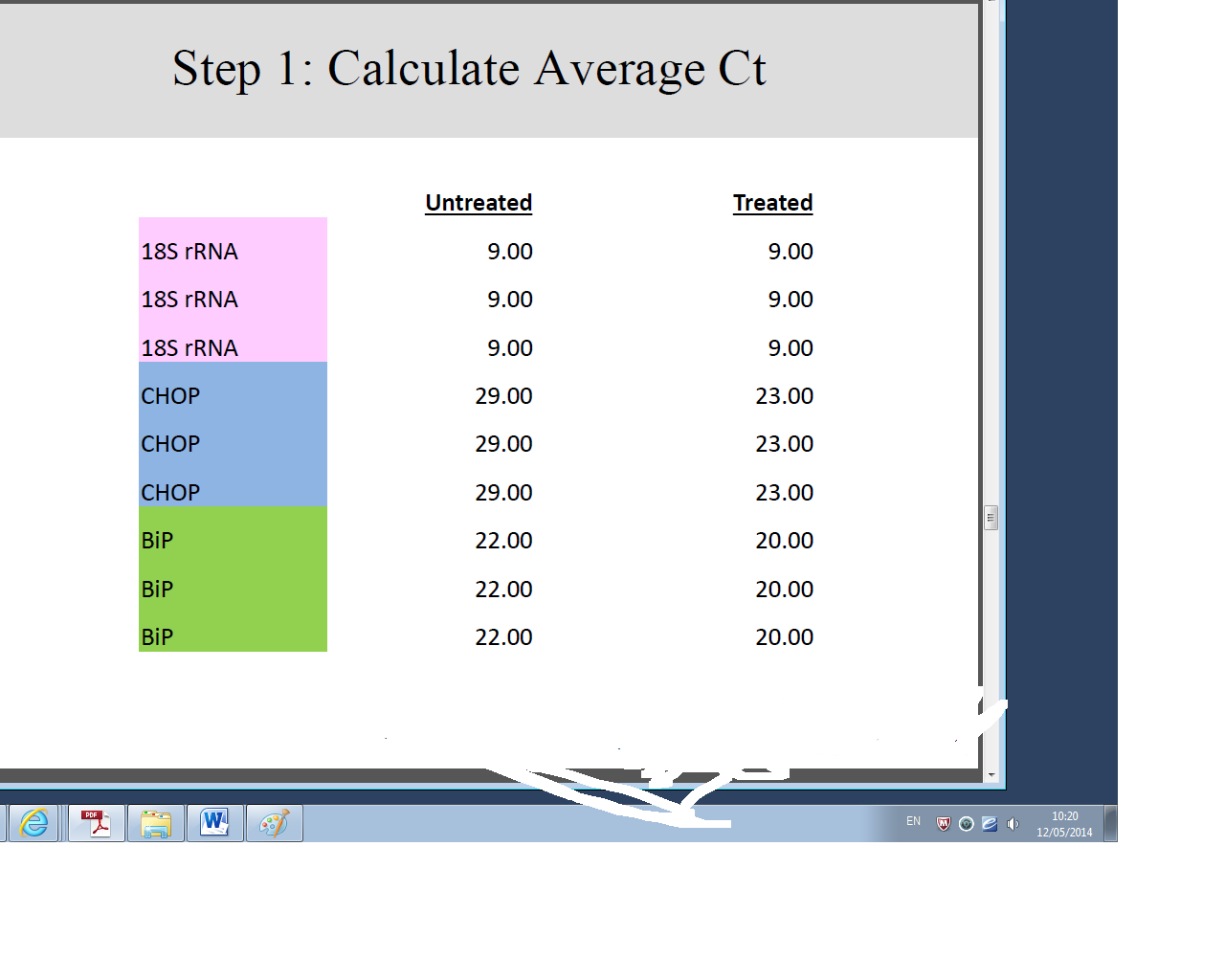
Differences between gene expressions of treated and untreated samples were established by a keen analysis of the data collected, in four well established steps.
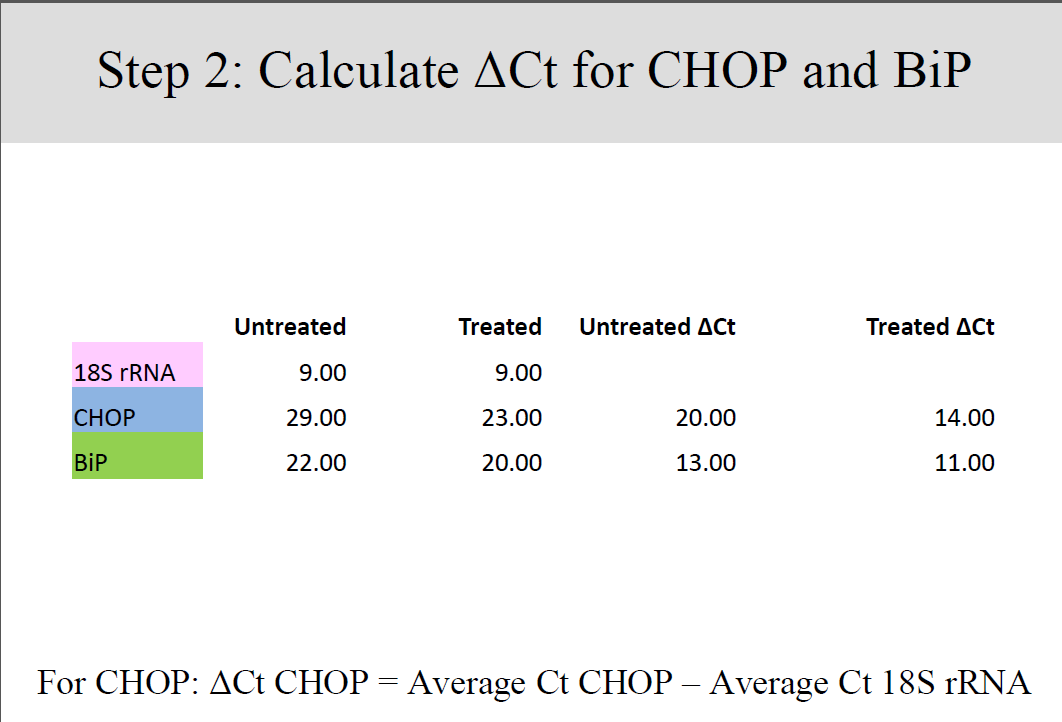
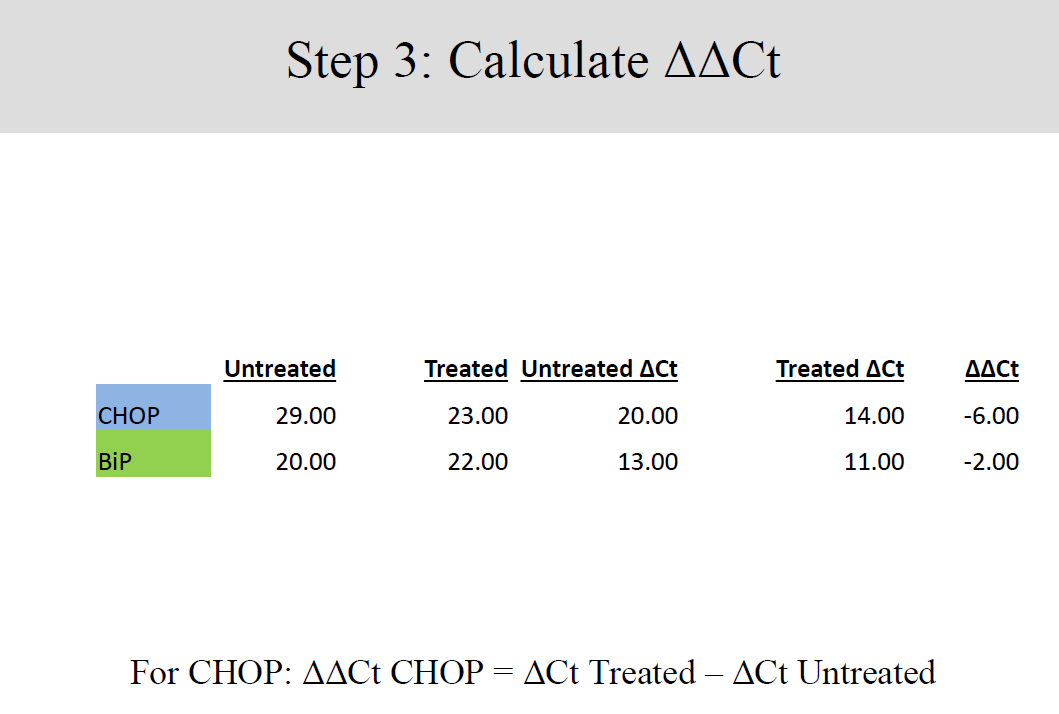
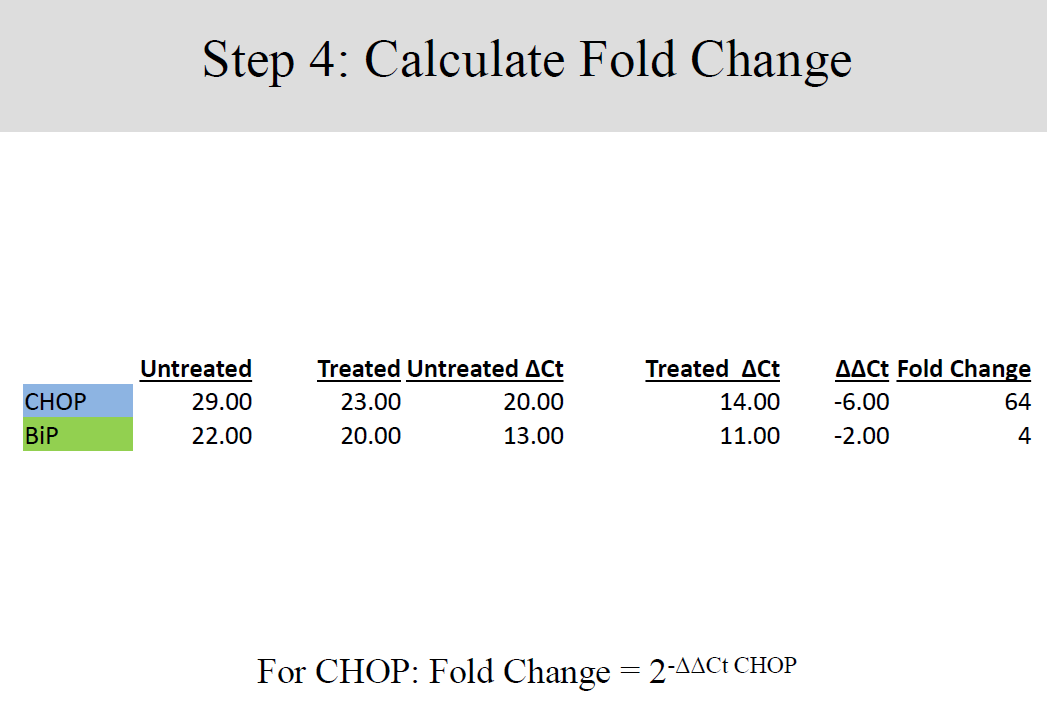
Conclusion and Discussion
These findings show a clear up regulation of the biological group treated with Tunicamycin as compared to the untreated group. Perturbation or stress of the endoplasmic reticulum during manipulation induces apoptosis thereby reducing the concentration of genetic materials in the collected samples. On the other hand, treatment by Tunicamycin ensures that endoplasmic reticulum stress is kept at bay hence apoptosis does not occur at an elevated rate (Tahmoorespur et al. 37). Therefore, cells treated with Tunicamycin have higher concentration of genetic material and exhibit higher levels of gene expression (Wyttenbach and Tolkovsky 1213).
These findings perhaps best explain why stress and perturbations in the eendoplasmic reticulum (ER) have repeatedly been cited as the source of neurodegenerative and ischemic diseases and conditions. Endoplasmic reticular stress incites the cells to engage a series of transcriptional response. This response is believed to be a result of protein unfolding and is called unfolded protein response (Wyttenbach and Tolkovsky 1213). The targets of unfolded protein response (UPR) include genetic molecules that characteristically act to minimize the effects of stress and perturbation. This they achieve via the inhibition of the destructive effects of aberrant endoplasmic reticular proteins. However, “severe and prolonged ER stress leads to the activation of an execution program by the cells” (Wyttenbach and Tolkovsky 1213). This stress-induced death in the ER results from the activation of intra-reticular caspase-12. This in turn activates caspase-3, which performs the execution role. Tunicamycin “is an inhibitor of protein glycosylation and acts in that regard to rapidly induce the expression of target genes” (Wyttenbach and Tolkovsky 1213).
Works Cited
Annis, Matthew, Naoufal Zamzami, Weijia Zhu, Linda Penn, Guido Kroemer, Brian Leber and David Andrews. “Endoplasmic reticulum localized Bcl-2 prevents apoptosis when redistribution of cytochrome c is a late event.” Oncogene 20.16 (2001):1939–1952. Print.
Bailey, Daniel, and Peter O’Hare. “TransmembranebZIP transcription factors in ER stress signaling and the unfolded protein response.” Antioxidants & redox signaling 9.12 (2007): 2305-2322. Print.
Ballester, María, Rubén Cordón and Josep Folch. “DAG Expression: High-Throughput Gene Expression Analysis of Real-Time PCR Data Using Standard Curves for Relative Quantification.” PLoS ONE 8.11 (2013):1-5. Print.
Ben-Ari, Elia. “The silence of the genes.” BioScience 49. 6(1999): 432. Print.
Fricker, Michael, Sofia Papadia, Giles Hardingham, Aviva Tolkovsky. “Implication of TAp73 in the p53-independent pathway of Puma induction and Puma-dependent apoptosis in primary cortical neurons.” Journal of Neurochemistry 114. 3 (2010): 772-783. Print.
Huang, Lin, Yaqiu Lin, Suyu Jin, Wei Liu, Yaou Xu and Yucai Zheng. “Alternative Splicing of Testis-Specific Lactate Dehydrogenase C Gene in Mammals and Pigeon.” Animal Biotechnology 23. 2(2012):114-123. Print.
Kroeger, None, None Messah, None Ahern, None Gee, None Joseph, None Matthes, None Yasumura, None Gorbatyuk, None Chiang and None LaVail. “Induction of endoplasmic reticulum stress genes, BiP and chop, in genetic and environmental models of retinal degeneration.” Investigative ophthalmology & visual science 53.12 (2012): 7590-7599. Print.
Livak, Kenneth, and Thomas Schmittgen. “Analysis of relative gene expression data using real-time quantitative PCR.” Methods 25.4 (2001): 402–408. Print.
Minematsu, Takeo, Takashi Harumi and Mitsuru Naito. “Quantitative genotyping by amplifying the polymorphic sequences of Pre-Melanosomal Protein (PMEL17) gene using real-time polymerase chain reaction in chickens.” British Poultry Science [Br Poult Sci] 49.5 (2008): 542-549. Print.
Shen, Shihao. “Widespread establishment and regulatory impact of Alu exons in human genes.” Crossmark 108.7 (2011): 761-767. Print.
Tahmoorespur, Mojtaba, Amir Taheri, Hamid Gholami and Maziar Ansary. “PCR-SSCP Variation of GH and STAT5A Genes and Their Association with Estimated Breeding Values of Growth Traits in Baluchi Sheep.” Animal Biotechnology 22.1 (2011): 37-43. Print.
Wyttenbach, Andreas and Aviva Tolkovsky. “The BH3-only protein Puma is both necessary and sufficient for neuronal apoptosis induced by DNA damage in sympathetic neurons.” Journal of Neurochemistry 96.5 (2006):1213-1226. Print.