Introduction
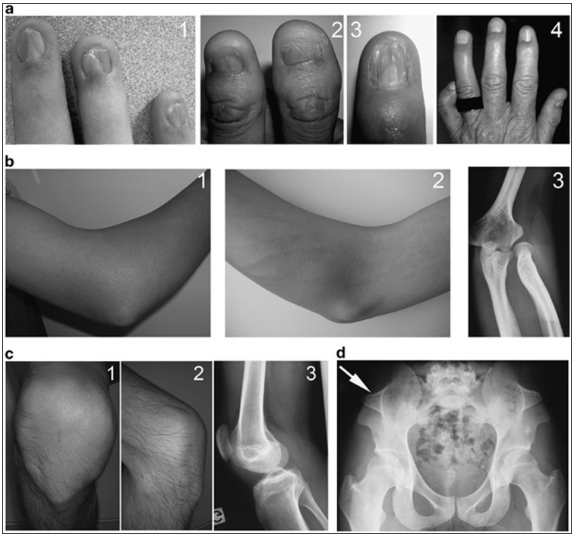
The field of genetic medicine has been considerably improved during the last centuries. Researchers use new technologies to approve their hypotheses and develop new approaches to develop their clinical and molecular studies. Each investigation is a possibility for people to comprehend the peculiarities of their genetic anomalies and health problems. One of the latest projects developed by Ghoumid et al. (2016) was published in European Journal of Human Genetics. The authors focused on Nail-Patella Syndrome (NPS) and the concept of a genetic heterogeneity. The article is full of clear explanations and definitions. NPS is defined as a rare autosomal dominant anomaly that is usually observed in human nails, elbows, and patella (see Figure 1).
The major aim of the study that the authors achieved was the necessity to check the validity of the hypothesis of a genetic heterogeneity in patients with Nail-Patella Syndrome. The authors took three stages in the study to achieve the goal. First, they described the phenotype and molecular data of a number of chosen patients and their relatives, who had NPS. Then, they identified possible rearrangements of LMX1B that was defined as the main disease-causing gene (Ghoumid et al. 2016). Finally, they discussed the possibility of a genetic heterogeneity in those patients.
Syndrome’s Description
Ghoumid et al. covered certain historical aspects of NPS and relied on several reports which helped to improve the clinical description of the syndrome (Roecherath 1951; Bongers et al. 2005; Sweeney et al. 2003). A range of clinical phenotypes was discovered, including nail dysplasia (anonychia, koilonychia, and triangular lunulae), elbow dysplasia, patellar dysplasia (hypoplasia, agenesis, and dislocation), iliac horns, intra-ocular hypertension, glaucoma, and peripheral neuropathies.
The authors explained that phenomenon of the syndrome regarding the existing variety of phenotypes. For example, renal anomalies were explained by asymptomatic proteinuria. However, the location of the mutation could vary, and the observable characteristics could vary as well defining its range, severity, and the development of abnormalities. In the article, the authors did not give clear explanations of the phenomenon, but focused on the enumeration of possible changes in human health and the conditions of nails, elbows, and patella.
It is possible to use additional sources and clarify that such factors as mutations in the gene, the possible effects of other illnesses, and even pregnancy cases could be used as the explanations for NPS (Álvarez-Martín et al. 2013).
As it has been already mentioned, LMX1B is the gene that causes the disease. The authors clarified the location of the gene (on the 9q34 chromosome) and its main components (eight exons in approximately 82 kb of genomic DNA) (Ghoumid et al. 2016). Besides, it was mentioned that LMX1B encoded an LIM-homeodomain transcription factor. That factor could be found in different developmental mechanisms. It is responsible for numerous pleiotropic manifestations that may be defined in the disease. In molecular biology, this factor is defined as a protein that is responsible for the regulations of the expressions of its target genes (Lee & Huang 2013).
Physical binding to the promoter of the genes is the main type of connection between the cells. In the study, Ghoumid et al. (2016) used two LIM domains in the protein (LIM-A and LIM-B) with two zinc fingers and one 60-amino acid homeodomain. Not much attention was paid to the possible impacts of mutations on the biological activity of the protein. Therefore, it is possible to address the studies developed by Sonqdej & Rao (2015) or Kamanu et al. (2012) about the possibility of mutations to influence the work of genes and to change the functions of molecular levels.
However, in Ghoumid et al. research, the homeodomain as defined as “highly conserved” meaning that DNA binding and transcriptional activation are the processes that cannot be neglected or misunderstood. This statement has been already proved by many researchers such as Hrycaj & Wellik (2016). It means that NPS is characterised by the functional and sequential conservation of the proteins.
However, the process of evolution may have its own consequences on the work of cells and the development of genes. The relevance of this statement remains to be crucial because future researchers and investigators could rely on the conclusions made by Ghoumid et al. (2016) including the place of evolution, the work of proteins, and the changes in cells due to the presence of LMX1B gene. The recognition of the homeodomain helps to create strong and relevant conditions for the following methods choice and the establishment of the criteria for the participants in the study under consideration.
In addition to the description of genes and their work, the authors of the article discussed several pathogenic mechanisms with the help of which the presence of dominant NPS was proved in humans. Those mechanisms included haploinsufficiency, dominant-negative effects, and gain-of-function effects. Haploinsufficiency is the medical phenomenon that introduces a diploid organism with one single functional gene copy and another gene’s copy that cannot work and produce the required protein because of mutation. Haploinsufficiency was discovered in patients, whose organisms had one of the LMX1B options or some partial deletions.
MRNA decayed, and transcriptional activity, as well as DNA-binding ability, could not work properly and turned out to be functionally diminished. As a result, the partial loss of LMX1B function was observed. In addition, it was hard to identify a genotype-phenotype correlation in extra-renal manifestations (Ghoumid et al. 2016). Therefore, the changes in renal involvement could also lead to such problem as dominant-negative and gain-of-function mutations.
Dominant-negative mutations are characterised by altered gene products and phenotypes that influence the functions of alleles. Gain-of-function mutations are able to change the gene products due to abnormal functions and the creation of new alleles. The description of the chosen phenotype depends on the variations that could happen to LMX1B, and the above-mentioned pathologies get the researchers prepared for different outcomes.
Families
In the article under analysis, the researchers paid much attention to the choice of the participants for the study. In general, 94 affected-with-NPS subjects were chosen. 55 major patients and 39 relatives of those patients were invited. The authors introduced some information about the participants in addition to those numbers. Gender differences, general geographical data, and the participants’ phenotypes were mentioned. That information was relevant and sufficient.
However, it seems that more facts and details had to be mentioned. For example, it was possible to clarify the connections between family members to underline the importance of genes and the role of generation in the study. Besides, the results of the study contained the information connected with the age of the patients. Therefore, it could be better and effective in case the researchers introduced each member of a family recognising their age. Such information is not hard to find and mention in the study to provide the reader and future researchers with a good background for their own work and understanding of the genetic heterogeneity hypothesis.
Methods and Results
Ghoumid et al. (2016) chose a molecular analysis as the main method of the study. On the one hand, it was expected that the authors addressed the data offered by Ensembl and relied on four protein coding transcripts that could be applied to LMX1B (Ensembl: gene: LMX1B n.d.). On the other hand, the authors mentioned that they used the nomenclature offered by GenBank and tried to compare their results with the results of the already collected data using the genetic analyser with the help of which the research could obtain reaction conditions and sequences on request (Ghoumid et al. 2016).
Taking into consideration the explanations and the identification of the numbers, it is possible to say that no problems or concerns should appear because the researcher established the boundaries before explaining the results and explained their goals clearly. The choice of transcripts usually depends on the splice variant the researchers are interested in. In this case, the subject was explained in details, and it was a good start in research. The coding sequence was properly agreed on between the past projects and the current study.
As soon as the choice of transcripts is explained, it is necessary to pay attention to another peculiarity of the article. The authors developed their molecular analysis on four different techniques and explained their choices. The basic technique was Sanger sequencing of exons. Its goal was to identify new variants of mutations and changes after the evaluation of exons and their flanking intronic regions.
In case no variants were discovered after that technique was used, it was suggested to use MLPA (multiplex ligation-dependant probe amplification) technique. It could help to identify possible duplications or deletions. Still, the authors thought that they might not find variants after those two techniques. Therefore, they tried to use another option in the form of array-based comparative genomic hybridisation that included log2 ratios, ADM-2 algorithms, and z-scores. Still, it was not the last technique the authors could rely on. In case the results of three above-mentioned techniques were negative, it was suggested to use genomic sequencing of whole LMX1B. With the help of that technique, the authors could explore the non-coding regions of the gene.
Each technique was used to support the previous one and provide the researchers with certain results that could be used in the analysis. Still, the main role was put on the results obtained from Sanger sequencing of exons. This sequencing technique remains to be the standard that is used to determine the primary nucleotide sequence (Hogge & Rajkovic 2014; Reisner 2014). It was performed in several simple steps: several peripheral blood samples were used to extract the DNA from them with the help of the EZ1 DNA blood kit, and eight exons were amplified by PCR and then sequenced on a special analyser.
At the end of that screening method, a molecular anomaly was discovered in 50 out of 55 families. It did not take much time to get the results and start analysing them. The same process and technique were developed by Edwards et al. (2015), who investigated the novel LMX1B mutation in a family. The choice of that technique was supported by many researchers and proved the appropriateness of the decisions made by the authors of the article.
Though Sanger sequencing was the main technique chosen by the researchers, a certain attention was paid to the technique of genomic sequencing of all LMX1B. The team decided to use that approach in case they were not able to find out the variant after three chosen methods. It was necessary to focus on the non-coding regions such as a promoter, 5’UTR, introns, and 3’UTR (Ghoumid et al. 2016). The decision was made after no molecular anomalies were defined in the eight exons of LMX1B in five families.
Besides, no considerable rearrangements were observed using the array-based comparative genomic hybridisation. It was discovered that 450 variants per patient were possible. Some of them were discovered in introns. In fact, some researchers already tried to discuss functional effects of a mutation in non-coding regions.
For example, Scacheri and Scacheri (2015) admitted that such functional regulatory elements as enhancers and insulators could undergo certain changes because of mutation and promote the development of congenital anomalies. Still, it is necessary to remember that non-coding functional elements may play different roles in regulating the expression of the gene. Therefore, DNA mutations may easily disrupt the functions of the gene even in non-coding regions.
Due to the location and the actual number of the variants, it was necessary to create some filters to look at the data obtained from the genomic sequence for all LMX1B. In the article, three main filters were discussed. They were the homozygous variants which could assume the autosomal dominant mode, the recurrent variants which could be identified in control samples, and the single nucleotide polymorphisms (Ghoumid et al. 2016). In fact, the choice of these filters creates a number of doubts.
On the one hand, the decisions turned out to be clear, and the authors used their system to explain how the genomic sequencing for all LMX1B technique worked. On the other hand, it was hard to identify the main criterion in regards to which the division was made. It was not always possible to divide all family members properly, and two variants with an uncertain significance were left. For example, it was possible to divide the results according to affected and unaffected individuals or use the VQSR filter (Variant, Quality, Score, and Recalibration) in order to be able to generate the errors and calculate the probabilities of every variant separately (Carson et al. 2014).
Findings
In modern studies, the researchers have access to different prediction tools, also known as in silico prediction tools. They are usually used to perform a bioinformatics analysis in a short period of time. Their main feature is the possibility to predict the pathogenicity of a variant defined in a result of any sequencing analysis. In the study under consideration, the researchers use Polyphen 2 that includes protein sequence and structure peculiarities and SIFT that explains sequence and evolutionary conservation (Ghoumid et al. 2016).
Polyphen 2 helps to predict the possible impact that may be caused by an amino acid substitution and change the structure or even the function of a human protein. SIFT is the tool that is based on sequence homology considered on the basis of multiple sequence alignments which help to predict the mutation pathogenicity. Regarding the main characteristics of the tools, the predictions and explanations are reliable. At the same, it is also possible to use PROVEAN tool software of which could help to focus on the impact of the biological function of the chosen protein thoroughly. Still, the choices of the researchers had enough grounds, and it is wrong to make attempts and improve their research. New methods and tools could be used in a new investigation only.
Some critiques and researchers may put the idea of two domain mutation being used for all identified variants under a question. Still, it is necessary to remember that, at the beginning of the study, the authors identified LIM and HD domains. The point was that the chosen protein comprised two types of domains. It could be wrong and inappropriate in case another number of domains or types of mutations were mentioned in the results. Therefore, mutations have to cluster in two domains at the same time, and the offered results covered the idea in a proper way.
Another interesting feature in the discussion of the findings was connected with the results of family 51. The results of the segregation analysis introduced a number of variants which were generated by next-generation sequencing. Besides, the authors paid their attention to microsatellite markers. They also admitted that no linkage to LMX1B locus was observed (Ghoumid et al. 2016). That statement could be proved by the genomic alteration of the chosen gene and the impossibility to connect it with any of family’s members. Therefore, the offered hypothesis of a genetic heterogeneity has been discussed before. It was hard to find the connection and develop similar ideas.
Summary
The article under analysis has a number of strong and weak aspects. Discussing its strong points, it is necessary to mention a powerful and informative background on Nail-Patella Syndrome and the organs or the parts of the body that undergo some changes when a person is diagnosed with this disease. Besides, the authors properly discussed different types of involvements that help to understand the functions and the structure of the chosen protein. For example, skeletal involvement (including patella, scapula, and elbow) is inherent to more than 89% of all participants. Nail involvement is another crucial factor that includes 88% of the patients. Renal assessments play an important role as well because approximately 80% of the patients had to deal with urinalysis and creatinine clearance measurements (Ghoumid et al. 2016).
Finally, the authors suggested to use some ophthalmological assessments to cover the possible problems with hypertension. Considering the weaknesses of the study, the sample and general information should be mentioned. The authors did not focus on discussing the peculiarities of their choices of the participants. They also could not give clear explanations why one prediction tool was chosen, and another tool was refused or omitted. In other words, the scientific validity and importance of the study are high indeed. Still, this project could be improved in the future in order to explain how people could deal with NPS and what changes in structure and function could be expected.
Reference List
Álvarez-Martín, N, Gamundi, MJ, Hernan, I, Carballo, M, Luis-Yanes, MI & García-Nieto, V 2013, ‘Nail-patella syndrome: a case with a de novo mutation in the LMX1B gene not previously described’, Nefrologia, vol. 33, no. 4, pp. 585-586.
Bongers, EM, Huysmans, FT, Levtchenko, E, de Rooy, JW, Blickman, JG, Admiral, RJ & Hyugen, PL, 2005, ‘Genotype-phenotype studies in nail-patella syndrome show that LMX1B mutation location is involved in the risk of developing nephropathy’, European Journal of Human Genetics, vol. 13, no. 8, pp. 935-946.
Carson, AR, Smith, EN, Matsui, H, Braekkan, SK, Jepsen, K, Hansen, JB & Frazer, KA 2014, ‘Effective filtering strategies to improve data quality from population-based whole exome sequencing studies’, BMC Bioinformatics, vol. 15, no. 125, pp. 1-15. Web.
Edwards, N, Rise, SJ, Raman, S, Hynes, AM, Stivastava, S, Moore, I, Al-Hamed, M, & Xu, Y 2015, ‘A novel LMX1B mutation in a family with end-stage renal disease of ‘unknown cause’, Clinical Kidney Journal, vol. 8, no. 1, pp. 113-119.
Ensembl: gene: LMX1B n.d. Web.
Ghoumid, J, Petit, F, Holder-Espinasse, M, Jourdain, AS, Guerra, J, Dieux-Coeslier, A, Porchet, N, Manouvrier-Hanu, S & Escande, F 2016, ‘Nail-Patella syndrome: clinical and molecular data in 55 families raising the hypothesis of a genetic heterogeneity’, European Journal of Human Genetics, vol. 24, no. 1, pp. 44-50.
Hogge, WA & Rajkovic, A 2014, Practical genetics for the ob-gyn, McGraw Hill Professional, Berkshire.
Hrycaj, SM & Wellik, DM 2016, ‘Hox genes and evolution’, F1000Research, vol. 5, no. 859. Web.
Kamanu, FK, Medvedeva, YA, Schaefer, U, Jankovic, BR, Archer, JA & Bajic, VB 2012, ‘Mutations and binding sites of human transcription factors’, Frontiers in Genetics, vol. 3, no. 100, pp. 1-6. Web.
Lee, C & Huang, CH 2013, ‘LASAGNA: a novel algorithm for transcription factor binding site alignment’, BMC Bioinformatics, vol. 14, no. 1, pp. 108-121. Web.
Reisner, H 2014, Pathology: a modern case study, McGraw Hill Professional, Berkshire.
Roecherath, W 1951, ‘Hereditary osteo-onychodysplasia’, Fortschritte auf dem Gebiete der Röntgenstrahlen, vol. 75, no. 6, pp. 700-712.
Scacheri, CA & Scacheri, PC 2015, ‘Mutations in the noncoding genome’, Current Opinion in Paediatrics, vol. 27, no. 6, pp. 659-664.
Sonqdej, N & Rao, AK 2015, ‘Hematopoietic transcription factor mutations and inherited platelet dysfunction’, F1000Prime Reports, vol. 7, no. 66. Web.
Sweeney, E, Fryer, A, Mountford, R, Green, A & McIntosh, I 2003, ‘Nail patella syndrome: a review of the phenotype aided by developmental biology’, Journal of Medical Genetics, vol. 40, no. 3, pp. 153-162.