Introduction
Molecular cloning is a set of methods in molecular biology that is used to obtain multiple copies of the target DNA fragment. This procedure includes several steps: preparation of cloning vector and a target DNA fragment, preparation of a recombinant DNA, and its insertion into a host organism. Target DNA fragment – it is usually a coding gene or a regulatory element. For molecular cloning procedures, it should be isolated, amplified, and purified. As a vector, bacterial plasmids and bacteriophages are used.
Bacterial transformation is a process of recombinant DNA (a cloning vector linked with a target DNA fragment) insertion into a host bacterial cells (1). Various methods could be used for bacterial transformation with plasmid vectors, including chemical treatment of cells with salts (in particular, CaCl2), heating the bacterial suspension, or electroporation. All the approaches have the same principle: after the treatment, small holes appear in the bacterial membrane, which makes the process of plasmid uptake easier (2).
The electroporation technique is widely used for bacterial cell transformation. It was stated that the efficiency of this process is up to 80% of survived transformed cells (3). For this procedure, the suspension of bacterial cells and recombinant DNA plasmids are subjected to brief electrical impulses. It results in small holes that appear in bacterial membranes. Plasmids could easily penetrate into bacterial cytosol through these holes (4).
After plasmid insertion, it is required to select transformed cells. Several approaches are used for this procedure. A recombinant vector contains genes that allow the identification and selection of transformed cells. It could be a gene of certain antibiotic resistance (1) or a Green Fluorescent Protein (GFP) gene. GFP is a protein from jellyfish Aequorea Victoria. It is responsible for glowing points of light around the margin of its umbrella (5).
In response to ultra-violet radiation, GFP produces green fluorescence emission. According to the technique, the GFP-coding gene introduced downstream of the promoter region of a target gene. Therefore, if cells produce GFP, they also produce target molecules. GFP gene expression can be easily detected under the ultra-violet radiation. Bacterial colonies that produce green fluorescence could be visually identified and isolated (6).
GFP plasmids (pGFPs) are widely used in molecular cloning techniques in biomedical researches. It is used for human cells transformation in cancer studies (7), animals (8), and bacterial (9) cells transformation. In biotechnology, molecular cloning with pGFP is widely used to insert genes of target protein production (enzymes, human antibodies, recombinant medicaments such as antibiotics and insulin, protein for vaccines production, and others) (10).
In this laboratory report, the process of bacterial cell transformation is described. This project is dedicated to recombinant plasmid construction and bacterial cells transformation. It includes several steps:
- Growing Escherichia coli bacteria with a plasmid with a gene of ampicillin resistance (pGFP) and a plasmid with a gene of Chloramphenicol resistance (pBCKS) and plasmids isolation.
- GFP gene from pGFP restriction and its insertion into pBCKS.
- Bacterial cells with recombinant pBCKS transformation.
- Transformed cells detection and selection.
Restriction enzymes treatment is used to isolate GFP fragment from the ampicillin vector. In the Chloramphenicol vector, GFP fragment occurs behind the Lac polymerase promoter region. In this case the plasmid sequence orientation and GFP gene expression are correct. The final step of the project is bacterial selection. Successfully transformed bacteria cells will be Chloramphenicol resistant and will produce green fluorescence emission with the UV light.
Aim
The aim of this project was to insert Green Fluorescent Protein (GFP) coding gene into pBSCK vector and to introduce this vector into E. coli bacteria cells.
Materials and Methods
All materials and procedures were explained in “ONPS1052” RMIT Gene Technologies 1 Practical notes 2017 by Mouradov (2).
Results
In the study, pGFP and pBCKS plasmids were used. Figure 1 shows the structure of plasmids.
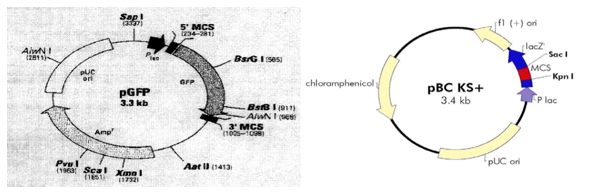
Both plasmids have the similar structure. The pGFP vector contains a selectable marker (the ampicillin resistance gene) and a visual marker (GFP). The pBCKS contains chloramphenicol resistance gene as a selectable marker. Both plasmids contained multiple cloning sites (MCS) with restriction sites for EcoRI and HindIII enzymes (2).These plasmids were inserted into competent E. coli strains. Liquid LB cultural media with ampicillin and chloramphenicol antibiotics were used for growing transformed E. coli. Bacteria cells numbers are shown in Table 1.
Table 1. E. coli with pGFP and pBCSK plasmids colonies count on LB medium containing ampicillin or chloramphenicol correspondingly.
The high number of transformed single E.coli colonies containing pGFP plasmid and pBCKS plasmids grow on LB medium containing either ampicillin antibiotic (pGFP ampicillin resistance) or Chloramphenicol antibiotic (pBCKS Chloramphenicol resistance). It could be stated that pGFP transformation was more efficient because the concentration of transformed cells with pGFP per 10 µl of a suspension was more than twice higher, and per 100 µl was around 1.6 times higher in comparison to pBCKS transformed cells’ concentration. The difference between 10 µl and 100 µl results could be considered as insignificant. In general, both processes of transformation were successful, and cells with target plasmids were obtained.
Extracted plasmids treated and non-treated with EcoRI and HindIII enzymes were analyzed by electrophoresis in agarose gel. The results are presented in figure 2. According to the results, digested pGFP contained non-restricted GFP plasmid (~2600bp) and a fragment of GFP gene (~800bp). Digested pBCKS contained non-restricted plasmid (~3100bp).
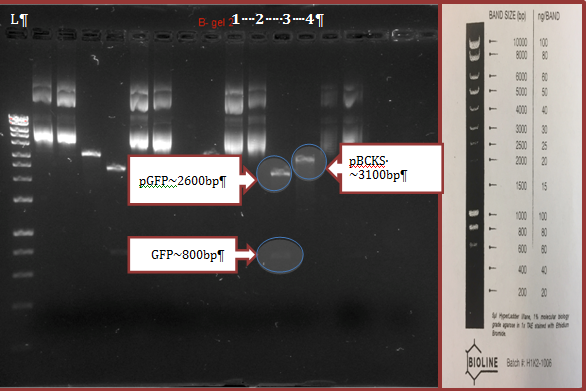
Group results (1-4 lines) of electrophoresis in agarose gel of extracted plasmids treated by digestive enzymes (EcoRI + HindIII) as well as non-treated plasmids. L= Ladder; lane number 1= pGFP; lane number 2= pBCKS; lane number 3= Digested pGFP; lane number 4= Digested pBCKS.
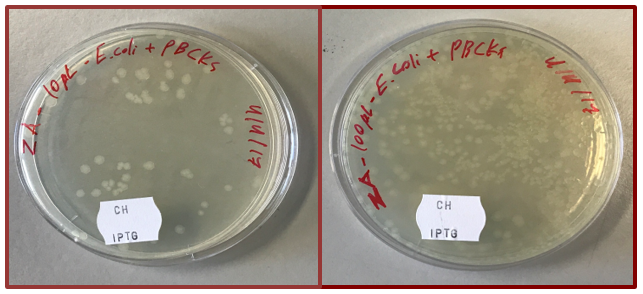
The procedure of ligation of two restricted plasmids was conducted. As a result, a new recombinant pBCKS plasmid contained BFP gene was expected to appear. On the last stage of the project, competent E. coli cells were transformed with the recombinant plasmid (ligation of the GFP with pBCKS backbone). 10 and 100 µl of transformed bacteria mixture were cultivated on a solid LB medium with chloramphenicol (Ch.) and isopropyl β-D-1-thiogalactopyranoside (IPTG) (fig. 3).
Only cells with introduced recombinant pBCKS plasmid were able to grow on this media. Chloramphenicol was used to select bacteria with a gene of resistance to this antibiotic which was a part of pBCKS. IPTG is a compound which induced the expression of genes which are controlled by Lac promoter. Therefore, only in presence of this compound, plasmid genes expression is possible (11).
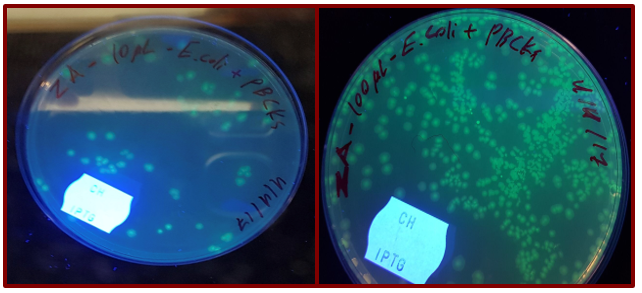
The presence of GFP gene was detected under the UV light. Under this condition, GFP started to immerse green fluorescence which could be easily detected by the naked eye (fig. 4).
Discussion
The initial transformation of E. coli using both types of plasmids, shown in Figure 1, was successfully conducted. The high number of single E.coli colonies was obtained on solid LB medium contained either ampicillin or chloramphenicol antibiotics as selection factors for E. coli containing pGFP and pBCKS plasmids, correspondingly. The results are shown in Table 1. After culturing of a few colonies in LB broth and incubating overnight, plasmids were then extracted from transformed E. coli cells and digested with HindIII and EcoRI restriction enzymes. The digested and non-digested plasmids were run on the electrophoresis in agarose gel. Results are shown in Figure 2.
In case of non-digested plasmids, pGFP plasmid (the lane number 1, Figure 2), two bands were obtained, while it was expected o get one band around 3.3Kb. For non-digested pBCKS plasmid (the lane number 2, Figure 2), results were the same. In case of digested pGFP plasmid in the lane number 3 (Figure 2), two bands around 2600bp and 800bp were observed. In the related to digested pBCKS lane number 4 (Figure 2), one band slightly over 3100bp was observed.
After completing a sub-cloning stage, 10 µl and 100 µl of transformed E. coli contained recombinant pBCKS plasmids were cultured on an LB plate with chloramphenicol and IPTG (Figure 3). After cultivation of 10 µl mixture, 43 single E. coli colonies were observed on a plate with selective medium. After cultivation of 100 µl, approximately 250 single E. coli colonies were observed. The green fluorescence emission was detected under UV light, as it is shown in Figure 4.
In this study, it was confirmed that the plasmids could be successfully extracted from transformed E. coli. More than one 3300bp and 3400bp bands were observed on the gel for extracted pGFP and pBCKS plasmids, respectively. This could be explained by the fact that plasmid DNA can exist in three conformations on the gel: supercoiled, open circular, and linear. Linear DNA migrates with the slowest speed, in comparison to other forms (12). Therefore, the lowest observed on the gel bands (Figure 2) are assumed to be supercoiled form of non-digested pGFP and PBCKS plasmids. In case of digested pGFP plasmid, GFP band was detected at 800bp as it was expected. It means that the target GFP gene fragment has been successfully restricted by enzymes (HindIII and EcoRI).
After sub-cloning the GFP fragment to pBCKS plasmids and cultivation E. coli on an LB plate contained chloramphenicol and IPTG, the correct orientation of GFP sequence behind the lac promoter sequence was examined by screening emission of green fluorescence from E. coli colonies under the UV light. Interestingly, high number of green fluorescence emission E. coli was observed on the selected media. It means that the ligation was conducted efficiently with the high rate of recombinant plasmid formation. After that, the process of transformation was also effective. Therefore, a high number of E. coli colonies with plasmids contained both chloramphenicol resistance genes and GFP genes was obtained in the study.
In general, this project demonstrated the efficiency of protocols of plasmids isolation, restriction, and legation and transformation of competent E. coli cells with recombinant plasmid. Effective methods of detection (growing on selective media contained antibiotics and visualization of green fluorescence under UB light) were tested. Besides, the effectiveness of plasmid restriction was tested using the electrophoresis in agarose gel.
References
- Rapley R, Whitehouse D, editors. Molecular biology and biotechnology. London: Royal Society of Chemistry; 2014.
- Mouradov A. RMIT gen technologies practical notes, 1st semester. Melbourne: School of Applied Sciences, RMIT University; 2017.
- Jordan CA, Neumann E, Sowers AE, editors. Electroporation and electrofusion in cell biology. Berlin: Springer Science & Business Media; 2013
- Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucl. Acids Res, 1988; 16: 6127-6145.
- Shimomura O, Johnson FH, Saiga Y, Cell J. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. Journal of Cellular and Comparative Physiology, 1962; 59: 223-239.
- Cseke LJ, Kirakosyan A, Kaufman PB, Westfall MV, editors. Handbook of molecular and cellular methods in biology and medicine. 3rd ed. Boca Raton: CRC Press; 2016.
- Gao J, Chang MT, Johnsen HC, Gao SP, Sylvester BE, Sumer SO, et al. (2017). 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Medicine, 2017; 9(1): 4-17.
- Karunakaran DKP, Kanadia R. In vivo and explant electroporation of morpholinos in the developing mouse retina. Morpholino Oligomers: Methods and Protocols, 2017; 1565: 215-227.
- Baty AM, Eastburn CC, Diwu Z, Techkarnjanaruk S, Goodman AE, Geesey GG. Differentiation of chitinase-active and non-chitinase-active subpopulations of a marine bacterium during chitin degradation. Appl. Environ. Microbiol., 2000; 66: 3566–3573.
- Lipps G. Plasmids: current research and future trends. New York: Horizon Scientific Press; 2008.
- Gene technologies. Practical manual. Melbourne: School of Applied Sciences, RMIT University; 2015.
- Swamy PPM. Laboratory manual on biotechnology. New Delhi: Rastogi Publications; 2008.