Introduction
The use of instrumental methods for the qualitative identification of microorganisms in the laboratory has increased in practical value. The number of life scenarios in which unknown bacteria or other pathogens are to be identified is large, so there is a need for a detailed study (Zhang et al., 2021). Qualitative identification techniques differ not only in terms of the procedure but also in the properties based on which microorganism distinction is made (Tankeshwar, 2022). The present work proposes to perform qualitative identification using the Gram staining method and then using the API 20 E Multitest.
The present laboratory work is structured around the consistent use of identification techniques that narrow down assumptions about the unknown microorganism. In particular, it is paramount to determine the nature of the bacterium by Gram classification, labeling it either Gram(+) or Gram(-) (Tripathi & Sapra, 2022). Gram staining is used to distinguish between two broad clusters of bacteria, which differ in the width of the peptidoglycan and the presence of an outer membrane within the bacterial envelope.
Thus, gram-positive bacteria have a wide outer layer of peptidoglycan, in contrast to Gram-negative bacteria, whose peptidoglycan is short and protected by an outer membrane. The presence of such a wide layer in the case of Gram-positive bacteria fixes crystal violet molecules, so the bacterium leaves a violet color after the staining procedure. If the bacterium is found to be Gram-negative, a parallel API 20 E Multitest (Tankeshwar, 2022) can be used to identify it further. This test runs up to 20 assays simultaneously for different metabolic properties of an unknown bacterium, and based on the results, the nature of the microorganism can be determined. This report describes the procedures and results of the laboratory study performed.
Materials and Methods
The following materials were used for the laboratory analysis:
- Blood agar plate.
- Unknown specimen “A”.
- Unknown specimen “B”.
- API 20 E Multitest plate: API 20 E test strips, incubator (24-48 hours at 35°C).
- Gram staining components: iodine, ethanol, crystal violet, safranin.
- Lab utensils: microscope, sterile loop.
The lab work was based on experiments that reduced the number of potential candidates for the unknown microorganism. The professor gave a plate with unknown specimens, “A” and “B,” which had to be identified. Both samples were placed on a blood agar plate, which is a nutrient medium with the addition of defibrinated blood. The bacteria were stained with crystal violet, and excess pigment was washed off with distilled water. An iodine solution was then added as a mordant, and after rinsing with water, ethanol was added as a decolorizing agent.
Contrast staining with safranin was used for differentiation because Gram(+) bacteria retained their original purple color, while Gram(-) bacteria stained with the reddish tint of safranin. Each of the “A” and “B” stained specimens was then subjected to microscopic observation to describe the morphology and location of the colonies. Depending on the sample type, they were subjected to different types of further identification assays. Specifically, Gram(+) was subjected to catalase and VP tests, and the API 20 E Multitest was used for Gram(-). The results were recorded on a data sheet and then used to qualitatively identify the unknown sample.
The API 20 E test is used to perform 20 parallel tests to evaluate the fusion capacity and enzymatic reactions of the sample in different media. The sample was transferred to each well of the API 20 E plate using a sterile loop and then the entire plate was incubated at 35°C for 24-48 hours.
Results
The initial qualitative identification revealed that sample “A” was Gram-negative and sample “B” was Gram-positive. On microscopic examination, as shown in Table 1, specimen “A” had the morphology of rods, and specimen “B” had cocci. It is noteworthy that the bacteria of sample “B” were linked in ordered chains, while the arrangement of microorganisms of “A” was random and irregular. Further independent research was conducted only on sample “A” (Fig. 1), which was subjected to the API 20 E Multitest.
Table 1. Colony and sample descriptions of side A and side B


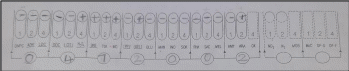
Figures 2 and 3 show the results of the API 20 E for sample “A,” displaying the positive and negative responses of the sample in each of the tests. As the results show, the sample gave positive reactions for the production of hydrogen sulfide, urease, tryptophan deaminase, tryptophanase for indole synthesis, gelatinase, and pentose sugar arabinose (Tankeshwar, 2022). However, the VP test for acetylmethylcarbinol production and the citrate test for carbon source gave adverse reactions.
Based on the results, the seven-digit sample code number was determined to be 0472002. Use of the API catalog shows that this code is characteristic of the Gram-negative bacterium Morganella morganii. As an additional check, the Motility Test gave a positive result for sample “A,” from which it follows that the bacterium is capable of free movement (Aryal, 2022). The result is confirmed: Morganella morganii is capable of independent movement by means of peritrichous flagella.
Discussion and Conclusions
The present laboratory work was devoted to the qualitative identification of an unknown bacterium using staining methods and microbiological tests. The results showed that the unknown bacterium was Gram-negative, capable of free movement, and produced hydrogen sulfide, urease, tryptophan deaminase, tryptophanase, gelatinase, and arabinose. Based on these data, the unknown specimen was classified as Morganella morganii. Clinically, M. morganii is the cause of urinary tract infections and wound infections and is active in patients with compromised immune systems (Bandy, 2020). Usually, the bacterium is found in the human intestine, so it is typically found in feces; therefore, if pathogenic, the commensal is transmitted through unwashed hands, direct fecal contact, and food contamination (Bandy, 2020).
Accordingly, the best prophylaxis against infection is the observance of sanitary rules and hygiene, as well as a common diagnosis in patients with urological abnormalities. When an infection has already occurred, treatment is implemented with antibiotics, including combined effects. Such antibiotic therapy may consist of the use of carbapenems (a broad class of drugs that inhibit bacterial wall synthesis), cephalosporins (a broad class of drugs that inhibit bacterial wall synthesis), and/or aminoglycosides (drugs that inhibit ribosome activity and disrupt protein synthesis).
This laboratory work has been successfully carried out, and only a few recommendations are suggested for future improvements. In particular, it would be necessary to take several samples of one unknown bacterium and retest them to guarantee a low margin of error. In a clinical trial setting, the accuracy of the results is critical, so creating erroneous results is unacceptable. No other changes are necessary because the work has satisfied the experimental objectives.
References
Aryal, S. (2022, August 10). Motility test – principle, procedure, uses and interpretation. Microbiology Info. Web.
Bandy, A. (2020). Ringing bells: Morganella morganii fights for recognition. Public Health, 182, 45-50. Web.
Tankeshwar, A. (2022). API 20E test system: Results and interpretations. Microbe Online. Web.
Tripathi, N., & Sapra, A. (2022). Gram staining. NIH. Web.
Zhang, T., Niu, Z., Wu, F., Chen, Z., Xu, J., Jiang, K., & Lai, Z. (2021). Qualitative and quantitative detection of surgical pathogenic microorganisms Escherichia coli and Staphylococcus aureus based on ddPCR system. Scientific Reports, 11(1), 1-10. Web.