Abstract
Fungi are widespread in the marine environment (Clement 1999). The marine species which have been identified number 600-1200 which constitutes only 1% of the total known fungi (Blomberg and Adler 1993). Scientists have raised the question as to why so few fungi have opted to live in places away from the marine environment to which they are suited from studies. It is also interesting to note that many fungi which are salt-tolerant have not yet started living in areas appropriate for their survival. These include the Aspergillus which is known as the soil organism and the spoilage agent. Some marine fungi possess certain characteristics which enable them to survive in the marine environment. Dendryphiella lives in the marine environment. It belongs to the group of higher fungi which has ascomycetes and deuteromycetes and is a deuteromycete.
Of these Dendryphiella salina and Arenaria have been studied well. The growth of a fungus in a marine environment depends on the “sporulation characteristics, the ability to utilize different carbon or nitrogen sources, pH and temperature growth profiles, and tolerance to a variety of inhibitors, including NaCl” (Edwards et al 1998). These characteristics form the basis of studies on fungi. This paper elaborates the living conditions of the marine fungi, the studies that have been done so far on them, and the possibilities for the future finally discussing the results of the study being done here.
Introduction
Sporulation or spore germination of marine fungi is a feature of the saline environment (Clipson and Hooley 1995). Each characteristic is gene-controlled. The chances of genes controlling the option to live in a marine environment are high. Osmotic mutants which have changed abilities to survive in saline environments now exist. Most of the fungi have not been studied well yet within the marine environment and are believed to be pathogens for other marine species. Studies of the mutants allow us to decide the characteristic features of organisms. Salt-tolerant clones have been produced through modern technologies. This has been done by isolating a salt-tolerant gene from the Aspergillus and inserting it in the salt-sensitive mutant (Clipson and Hooley 1995).
The largest habitat of fungi is found in decaying vegetation close to the coastal areas with climates of temperate and tropical nature (Jones 1988). They are free-living or associated with the organic matter biodegradation. The salt concentration in the seawater at half molar is the deciding factor for life in the marine environment. The salt should not be toxic to the metabolic processes in the fungal cell and should be such that water stress is kept minimal. Evolutionary adaptation has probably assisted in the marine fungi taking in high concentrations of solutes to ensure osmosis of water into the fungi. However, the only salt available, sodium chloride, is toxic. Details of this adaptation are still not known (Clipson and Hooley 1995).
Dendryphiella
Dendryphiella salina happens to be different in that its physiology has been studied. Dendryphiella is a filamentous marine hyphomycete (Clipson and Jennings 1992). Synthesis of polyols and amino compounds that are compatible solutes helps to maintain the hyphal water potential at a lower level than the surrounding environment (Wethered, Metcalf & Jennings 1985). These compatible solutes are named thus as they allow the inflow of water into the cell and are not toxic. The control of the low concentration of ions in the cytoplasm is by way of the plasma membranes. The sodium and chloride ions across the plasma membranes in conjunction with membrane proteins. Clipson et al (1990) have studied the accumulation of ions. Other organisms which have walled cells use the same mechanism with some modifications.
Electron micrographs show pictures of walled cells with vacuoles and large volume of cytoplasm. This is in contrast with the cells of algae and higher plants which are mostly vacuoles. Vacuoles allow the taking up of more salt which can remain unharmed in them. When the environment is rich with organic substrates, marine fungi become organisms of decay. Synthesis of organic compatible solutes is a better strategy than the gross uptake of ions into the vacuoles (Clipson & Hooley 1995). Due to seawater salinities, 80% of the cytosolic volume is formed by the polyols and amino compounds (Clipson et al 1990).
Studies have recently isolated clones of salt tolerance determinants (Stanley, Hooley & Clipson 1995). Other studies have concentrated on the bioactive metabolites to identify the role of the organism in the carbon cycle in the sea (Grant and Rhodes 1992). The differences of this organism with the D.arenaria which is somewhat similar have been studied (Pugh and Nicot 1964). Kohlmeyer & Volkmann-Kohlmeyer, 1991 have found that the D.salina has longer conidia and more septa.
The vegetative hyphae of Dendryphiella have adapted to the salinity. Another study found that salt tolerance has been due to the water potentials. The mediation is believed to be through the synthesis of polyols and amino-N compounds. Salt tolerance is based on this factor and the accumulation of salt and water from the external environment. Ions however are not increased to the extent that would cause toxicity in the cytoplasm or cause hindrances in the metabolic processes (Clipson and Jennings 1990). Genetic studies are not advanced enough so the genetic control of the salt tolerance is not possible yet. The biotechnological potential of marine fungi has not been fully tapped due to the poor development of methodologies. Genetic cloning is not yet reliable enough for antibiotic or enzyme production from marine fungi. Dendryphiella salina does not have a sexual stage so genetic analysis seems impossible. A transformation protocol has not been planned and mutants have not been collected. The base upon which cloning vectors are formed (natural plasmids) has not been found.
Differences between the D.arenaria and D.salina (Edwards et al 2004)
Spore characteristics were not different at a ph of 6.5 and temperature of 25° whether observed at 7 days or 4 months (Edwards et al 2004). Exposure to varying salt concentrations showed a lesser growth of both at higher salt concentrations. The D. salina showed longer conidia and more septa than the D.arenaria. Broader conidia were seen in D Arenaria. The sodium chloride had some effect on the conidial length. Many variations was not seen as described by (Pugh and Nicot 1964).
Seawater salinity allowed the optimal growth of both varieties on solid media but the salt-free samples showed even better growth. One D.salina sample showed significantly less salt tolerance. Potassium chloride showed a less inhibited growth. However a combination of potassium and sodium chloride (salt) inhibited sporulation at lower temperatures but appeared to allow vegetative growth. Lithium chloride was toxic to all isolates and inhibited colony growth above 1M concentrations. The varying responses to salt concentrations could be attributed to the compartmentalization of ions or the induction of compatible solute synthesis (Clipson & Jennings 1992 cited in Edwards et al 2004).
The growth profiles and colony extensions were good in all the isolates at ph 6-9 (Edwards et al, 2004). D.salina showed an optimum of 5-7 though it was first seen in Galway Bay, Ireland where the ph is 6.5. This may reflect the adaptation to the alkalinity of the marine environment with ph 7.8. Some of the D.salina isolates showed good colony extension rates at 10°. The D. Arenaria showed good growth in a broader range of temperature even though of tropical origin. The dendryphiella species could be cosmopolitan and exist anywhere, temperature not being a determinant (Curran 1980 cited in Edwards et al 2004). Dendryphiella has the potential to exploit many nitrogen sources. This property facilitates its function as a saprotroph. Growth was vigorous when a vitamin supplement was added. The fluffy growth with profuse aerial mycelium was not seen in the arginine and ammonium as sources of nitrogen. Antibiotic hygromycin inhibited the growth in all the nitrogen sources. The resistance to hygromycin may be taken as a marker in the selection of a transformation system for the Dendryphiella (Edwards et al 1998).
Salt tolerance, the ability to grow in alkaline ph, the wide range of temperature that growth is possible in and the ability to grow in a variety of nitrogen sources with minimum requirements are important characteristics of a marine fungus. The exaggerated salinity tolerance of D. species at high temperatures suggests the possibility of adaptation in evaporating tidal pools exposed to the sun. The features of the D. salina and Arenaria confirm that marine fungi need certain properties which enable them to survive in the marine environment (Edwards et al 1998).This also helps to explain the inability of certain terrestrial fungi which are osmotically tolerant but still unable to live in the marine environment. Stages in their life cycles may not accept certain conditions of the environment. Aspergillus nidulans have been found to grow well in solid media but cannot survive in liquid media where just a small quantity of salt can inhibit growth (Cox et al 1995 cited in Edwards et al 2004).
Fungal genetics
The subject of fungal genetics was first studied in the 1940s when fungi began to be considered ideal for studying the nature of the genes (Hooley & Whitehead 2006). The one-one enzyme hypothesis was volunteered by the researchers Shear and Dodge. In the biochemical pathways, the stepwise transition from pre-cursor molecules to the products was studied; each step was controlled by a single gene encoding a single enzyme. This was made possible by the nutritionally-deficient mutants (Hooley and Whitehead 2006). Fungal genetics has been done only on a few terrestrial ascomycetes like the yeasts Saccharomyces cerevisiae Hansen and Schizosaccharomyces pombe Lindner. Marine fungi are those that can complete their life cycles in the marine environment (Kohlmeyer and Volkmann-Kohlmeyer 2003). Dendryphiella is a good physiological model for the study of genetics.
Ultra-violet light is a nonionizing radiation that affects marine fungi. The UVB light is that portion that reaches the Earth as the ozone layer is unable to halt UVB rays due to its depletion. DNA may be damaged by the UVB radiation (Magan 2007). The increased melanin content of the cell walls of the marine fungi absorbs the UVB radiation and prevents damage to the DNA. Radiation resistance may occur due to the DNA repair mechanisms (Magan 2007).
Classical genetics
The isolation and properties of mutants are involved in classical genetics. Defects or mutation in specific genes in relation to the changed physiological feature or developmental process is identified. Density-gradient centrifugation is used to isolate the mutants altered by glycerol metabolism (Hooley &Whitehead 2006).
Materials and Methods
Mutations have been used to define genes and after study of recombination and linkage behavior has helped to map the genes to the linkage in Aspergillus nidulans which has been studied in great detail. More powerful and better economical methods of recombinant DNA technology in the 1970s paved the way to more mutants and more studies with sure outcomes
Collection of fungi
The collection of fungi can be done through SCUBA and snorkeling.
Chromosome analysis
Conventional microscopy was insufficient to study the extremely small chromosomes of the fungi. Pulsed field gel electrophoresis causes the separation of chromosomes as bands in an electric field where the current is changed regularly (Hooley &Whitehead 2006). Corredor et al (2003) studied D. hansenii strains using this method. Four to ten bands of differing sizes were identified within the members of the genus. Chromosomal length polymorphism was found in the D.hansenii strains.
Cloning
Recombinant DNA technique in which genes are physically isolated in vitro is used for cloning. Identical copies are rapidly produced in sufficient quantity for studying the features like DNA sequencing. Transformation, complementation and hybridization are the techniques for cloning. Twenty-one isolates representing nine genera of marine filamentous fungi were screened by Baisden and Cooney (1996). Evidence of plasmids was found only in one. The transforming DNA is usually integrated into the host genome. This increases the stability of the transformant. In complementation, a gene library of the organism is made initially. Then the cloning is done by complementation of an altered phenotype. A host is selected from the gene library. Gene function can be tested directly by complementation or by restoring a normal type.
Clement e al developed a system whereby a D.salina gene library was transformed into A.nidulans with a salt sensitivity mutation (1999). Many potential clones were identified. A variety of genes can be cloned from a species using the polymerase chain reaction in short DNA sequences (Bridge 2002). This method is hybridization. It has been used for cloning genes encoding sodium pumps from D.Hansenii(Almagro et al 2001).
The salt or osmotic sensitivity mutations help to identify salt tolerance determinants from D.salina in A nidulans. Chlorneb resistance is increased along with the increased sensitivity to many osmotic agents through a phenotype chlA10 (Chabani and Grindle 1990). Another phenotype sltA1 produces a more specific sensitivity to certain inorganic salts (Spathas 1978).
An arginine-sensitive phenotype is also present (Clement et al 1994). This causes the host to have an abnormal arginase activity and thereby could be an allele of agaA (Clement et al 1996).
Co-transformation of the D.salina genomes made in lambda EMBL3 into the A.nidulans host which carried the sltA1 salt sensitivity mutation in the presence of the selectable marker for transformation pyrG89 has been done. The transformation scene was the growth in pyrimidine media when the previous property of the pyrG89 was the auxotrophy. It was able to identify D.salina clones which increased the salt tolerance of their host. Lambda packaging could separate some clones
into E Coli (Yelton et al 1985). This was for assessing the ability to complement sltA1 by retransformation into the original pyrG89 sltA1 A nidulans host (Stanley et al 1995). The transformation indicates that it is not mutational rearrangement that has conferred the new properties of salt tolerance but the incorporation of the incoming DNA at a locus on the chromosome. The packaged molecules can be introduced onto Luria Broth agar plates having ampicillin to isolate strains of Dendryphiella which can act as anti-microbials.
Molecular methods
Molecular methods are also used to identify isolates and the taxonomical relationships. Fourteen assumed structural loci of Dendryphiella were examined for genetic variation through isoenzyme profiles (Michaelis et al 1987). A numerical taxonomic approach was adopted by Edwards et al to compare isolates of D. salina and D. Arenaria. These species showed little variation in the absence of sexual reproduction.
Fungal isolates and growth
Isolates of D.salina and D.aneraria can be taken from already available fungal cultures available at research laboratories and established as single conidial isolates (Edwards et al 2004). Both were grown on cornmeal agar and compared following the method of Kohlmeyer and Volkman-Kohlmeyer (1991). Microscopic examination of colony growth by using 100 conidia from each isolate was performed. (Edwards et al 1998). Spores were transferred from 7-14 day cultures onto test plates. The ph of the agar was 7 and incubation was done at 25°C. After 120 hours, the colony diameters were measured in 3 different directions and another similar measurement was made after 40 hours. The growth or extension of the colonies during the interval period was then calculated. After 120 hours, spores were recognized by the green pigment visible to the eye in the mycelium.
For assessing the ph growth profiles, corn meal agar plates were arranged from ph 5-9. For determining temperature growth profiles, corn meal agar plates at ph 7 were incubated at temperatures between 10° and 30°. Tolerance to salt was determined by growth upon the cornmeal agar containing many salts of 0-1M. Tolerance to antibiotic Hygromycin was done by adding its sterile aliquots to the agar in the molten form. Nitrogen source utilization was done by inoculating the spores onto A.nidulans minimal medium (Clutterbuck 1974).
DNA fingerprints
Species-specific DNA fingerprints allowed the assessment of fungal diversity in the marine environment (Hooley & Whitehead 2006). Dendryphiella isolates from all parts of the world now use a new technique to examine variation: BIOLOG phenotype microarrays (Dela Cruz et al 2006). Variations of these techniques are frequently being used. Obligate and facultative marine fungi are defined using molecular techniques. All the DNA of an organism is cloned and sequenced in genome projects. Such information is organized into contigs or overlapping sections which are arranged in the specific chromosomes (Hooley & Whitehead 2006). The procedure of comparing the gene sets and chromosome positions is known as comparative genomics. Of the organisms suitable for genomic studies, D salina has emerged as a suitable candidate. The genomic approach is now widely applied to the microbial and metazoan communities of marine life (Wilson et al 2005).
Despite newer technologies, classical methods of ion sink (Clipson &Jennings 1992) are still good to isolate and study the osmotic response of cell wall mutants in D. salina. Stable morphological mutants of D.salina may be prepared by using mutagens like MNNG, ultra-violet light, and nitrous acid, influencing the spore characteristics and colony characteristics. Stress responses in the sea produce metabolites that may influence the production after a lag phase where no growth is seen. Kim et al (2003) claim that the world of new antimicrobials may be revealed with the molecular approach.
The biotechnology industry is very interested in the Dendryphiella DNA sequences and the encoded haloperoxidase and protease enzymes (Hooley & Whitehead 2006). D.salina is considered an imperfect marine species with little genetic variation. Studies are being done to predict the ability of the D.salina to withstand the climatic changes that may alter the marine environment. The increased fragility of the marine ecosystem makes it all the more important to continue explorations into marine life.
Salt tolerance has been examined on Dendryphiella salina on the host Aspergillus nidulans. This host is commonly found in temperate and subtropical regions. The high salt tolerance is seen in the vegetative states of A.nidulans(Tresner & Hayes, 1971). Aspergillus cannot be considered a marine fungus as its life cycle is not completely in the marine environment (Kohlmeyer & Volkmann-Kohlmeyer 1991). A. nidulans has vegetative forms in the marine environment. It is believed that sea fan has declined in population due to the A.nidulans. A.nidulans is a well-developed genetic tool, has efficient protocol transformation, an elaborate linkage map and a large number of mutants.
Results
Discussion
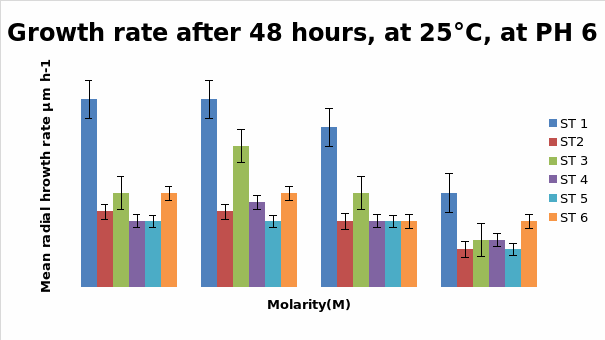
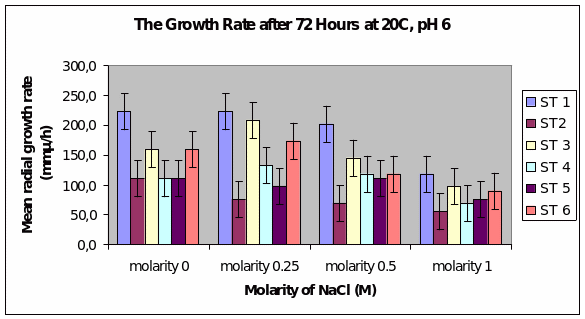
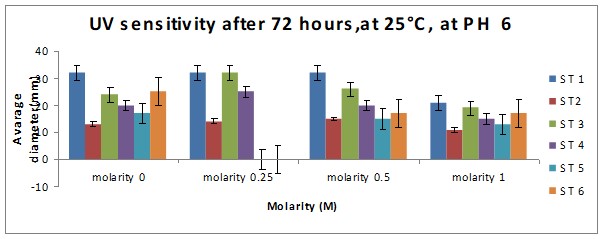
Bar Chart 1 All the isolates of D.arenaria, grown on malt extract and agar at 25°C at ph 6 with standard error being ± 1, showed the maximum mean radial growth rate at lower salt concentrations below 0.5M. Strain 1 showed exceptionally good growth in the salt-free and lowest concentration of salt at 0.25M, mean radial growth rate being more than 2000 microM/h. It showed more profuse growth than the other strains in all the salt concentrations tested. Strain 3 showed a special spurt of growth at 0.25 M, more than in the salt-free sample in which all the other stains were showing maximum growth. All the strains exhibited only about half their maximum growth in the highest salt concentration of 1M.
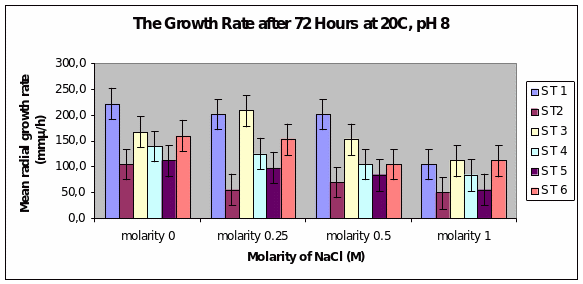
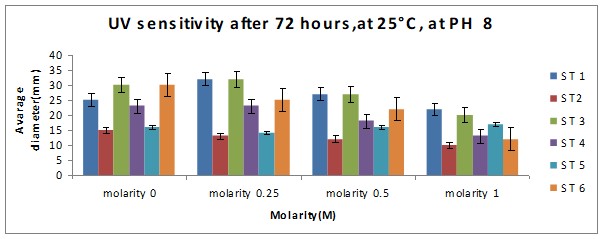
Bar chart 2 When ph was shifted to 8 or alkaline medium, the growth showed practically no difference from the pattern seen with ph 6. The salt concentration was again the factor that influenced the growth rate.
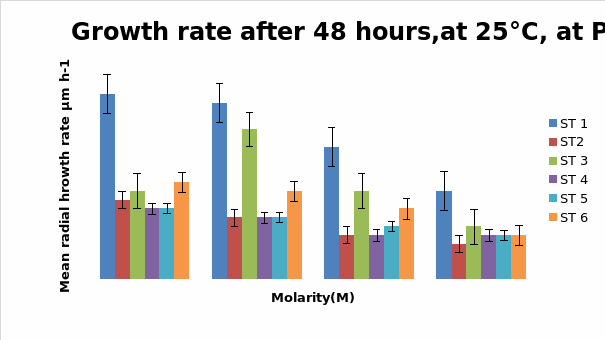
Bar chart 3 The change in ph had not affected the growth pattern seen in ph 8 above.
Bar chart 4: The chart showed that Strain 1 has had its growth slightly inhibited by the UV light in the salt-free sample. Strain 1 had accelerated growth in the low salt concentration sample of 0.25M. Strains 3 and 6 had excellent growth in the salt-free sample. The growth of Strain 3 was enhanced in the presence of a low salt concentration at 0.25M. while Strain 6 had its growth inhibited with lesser growth in the increasing concentrations. In the higher salt concentrations of 0.5M and 1M all strains showed decreased growth
Bar chart 5 After 72 hours at ph 8 at 20°C, Strain 1 showed the maximum mean radial growth among the strains at 2250 microM/h in the salt-free concentration. This was the same picture seen in the 0.25 concentration. Strain 3 showed a spurt of growth again in the 0.25 M. Strain 1 maintained its growth in the concentration of 0.5M but was inhibited in the 1M sample. All strains as previously had inhibited growth in the samples with highest salt concentration. The growth appeared similar to that with ph 6.
Bar chart 6 Strain 1 showed a better growth in ph 6 than in ph 8. Strain 3 showed a better growth in the low salt concentration than in the salt-free sample. All strains had their growth decreased by the higher salt concentrations.
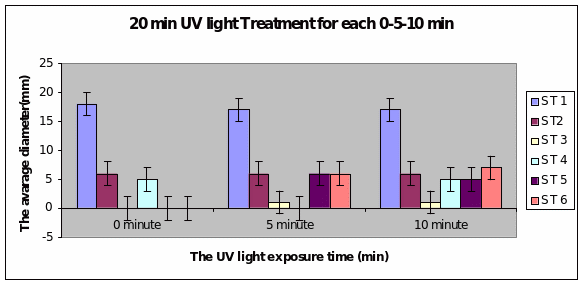
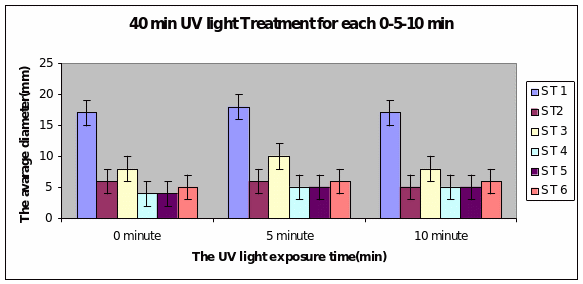
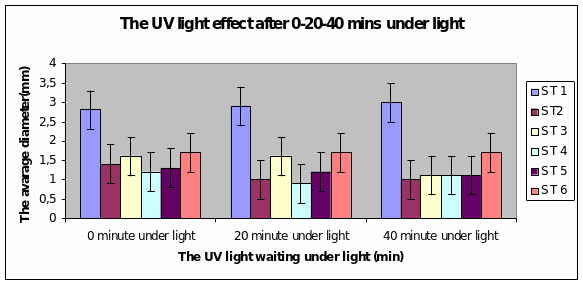
Bar charts 7 and 8 In both the 20min and 40 min UV, Strain 1 had no problem in growth. Strains 2 and 4 also started growing from zero hours in the 20 min experiment. The other strains showed growth after 10 minutes of the start of experiment. Strain 3 showed minimal growth even after 10 minutes in the 20 min UV exposure. However all showed growth from the start of experiment itself in the 40 min UV exposure
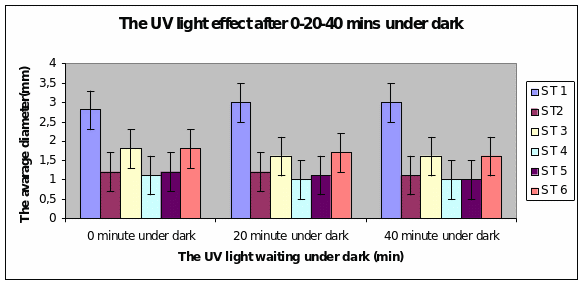
Bar charts 9 and 10 Strain 1 grew the best after 40 minutes under UV light. The same level of growth was seen in the dark too. The other strains showed growth from the start of the experiment and just maintained the same level even after 40 minutes of light. This was the same result even in the dark.
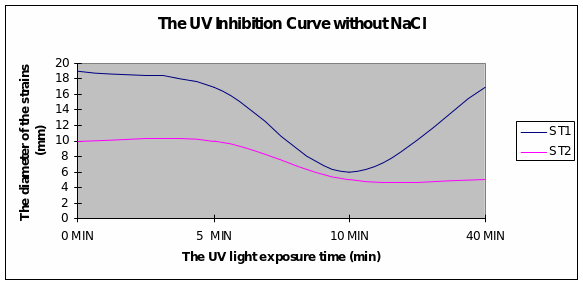
Graph 1 Strain 1 was inhibited after 10minutes of exposure to UV light. Strain 2 had a more uniform growth from the beginning.
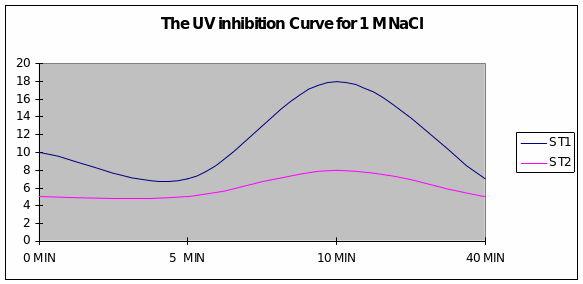
Graph 2 When the exposure to UV light was in the presence of sodium chloride of 1M concentration, a slight inhibition of growth after 5 minutes was overcome just after 5 minutes and reached maximum levels for Strain 1. Strain 2 had a uniform growth.
This study used malt extract and agar while Edwards’ (2004) study used corn agar meal.
The ph profiles were tested between 6 and 9 in Edwards’ study while in this study only two phs were used at 6 and 8. However the range was almost the same. The results of this study corroborated the evidence found in Edwards’ study (2004). Ph made no difference in the growth of the 6 organisms of D.arenaria.
Dendryphiella Arenaria Strain 1 appeared to be the best growing organism among the six. It could be described as the most salt-tolerant organism of the 6 organisms as it still had the best growth in the 1M concentration of salt even though its growth had reduced when compared to the salt-free sample in which profuse growth was seen. In almost all the conditions of ph, UV light radiation sensitivity, and salt concentrations it was found to have fairly profuse growth when compared to the others. All the other strains showed somewhat similar growth and their growths were reduced by the rising salt concentrations. Strain 3 showed surprising growth spurts in the low salt concentration samples. Whether this meant that nonsaline water was the best for this strain or whether to understand that this strain grew well in freshwater or whether it could grow in both fresh water and seawater was not clearly understood However its growth extent was nearly as much as Strain 1 in most of the samples.
Marine fungi are yet to be studied totally. They can be important pathogens of other marine species. The role of Dendryphiella as a saprotroph in removing decaying vegetation is significant to the marine environment. Further studies are necessary to identify means of stopping pathogenesis and improving the saprotroph function in the marine environment.
References
Almagro, A.et al, 2001. Cloning and expression of two genes coding for sodium pumps in the salt tolerant yeast Debaryomyces hansenii. Journal of Bacteriology,183, p. 3251–3255.
Baisden, C.M.& Cooney, J.J., 1996. Screening marine fungi for plasmids and characterization of a linear mitochondrial plasmid in a Lulworthia sp. Mycologia, 88, p. 350-357
Blomberg, A. & Adler, L. (1993) Tolerance of fungi to NaCl, in ‘Stress tolerance in fungi’, 209-232, Ed. D.H. Jennings. Marcel Dekker.
Bridge, P., 2002. The history and application of molecular mycology. The Mycologist 16, p. 90–99.
Chabani, A. W. & Grindle, M. 1990. Isolation, characterization and genetic analysis of mutants of Aspergillus nidulans resistant to the herbicide dichlobenil. Mycological Research 94, p.523-528.
Clement, D.J. et al, 1999. Complementation cloning of salt tolerance determinants from the marine hyphomycete Dendryphiella salina in Aspergillus nidulans Mycol. Res. 103 (10), p. 1252-1258 Printed in the United Kingdom
Clement, D. J., Attwell, N. A., Stanley, M. S., Fincham, D. A., Clipson, N. J. W.& Hooley, P., 1996 Evidence for sltA1 as a salt sensitive allele of the arginase gene (agaA) in the ascomycete Aspergillus nidulans. Current Genetics 29, p.462-467.
Clipson, N. J. W. & Jennings, D. H., 1990. The role of K+ and Na+ in the generation of the osmotic potential of the marine fungus Dendryphiella salina. Mycological Research 94, p. 1017-1022.
Clipson, N. J. W., Hajibagheri, M. A. & Jennings, D. H., 1990. X-ray microanalysis of the marine fungus Dendryphiella salina at different salinities. Journal of Experimental Botany 41, p.199-202.
Clipson, N.J.W. & Jennings, D.H., 1992, Dendryphiella salina and Debaryomyces hansenii:models for ecophysical adaptation to salinity by fungi that grow in the sea. Canadian Journal of Botany, 70, p. 2097–2105.
Clipson,N. and Hooley, P., 1995. Salt tolerance strategies in marine fungi Mycologist Volume 9, Part 1, P. 3-5
Corredor, M., Davila, A.M., Casaregola, S., Gaillardin, C., 2003. Chromosomal polymorphism in the yeast species Debaryomyces hansenii. Antonie Van Leeuwenhoek, 84, p. 81-88
Cox, P. W., Clipson, N. J. W., Hooley, P. & Thomas, C. R.., (1995). Effects of growth medium solute concentration upon the spore germination and colony growth characteristics of Aspergillus nidulans. Proceedings of the Institution of Chemical Engineers Research Event, Edinburgh 2, p. 986-988.
Clutterbuck, A. J., 1974 Aspergillus nidulans. In Handbook of Genetics, vol. 1, p. 447-510. Plenum Press : NY.
Dela Cruz, T.E. et al, 2006. Carbon source utilization by the marine Dendryphiella species D. arenaria and D. salina. FEMS Microbiology Ecology, in press.
Edwards, J. et al 1998, A comparative physiological and morphological study of Dendryphiella salina and D. arenaria in relation to adaptation to life in the sea. Mycol. Res. 102 (10), p.1198-1112 Printed in the United Kingdom
Grant, W. D. & Rhodes, L. L., 1992. Cell-bound and extracellular laminarinase activity in Dendryphiella salina and five other marine fungi. Botanica Marina 35, p. 503-511.
Hooley, P. and Whitehead, M. 2006. The genetics and molecular biology of marine fungi. Mycologist, 20, p. 144-1511, Sciencedirect.
Jones, E.B.G. (1988) Do fungi occur in the sea? Mycologist 2: 150-157.
Kim, C.F. et al, 2003. Cloning and expression analysis of the pcbAB-pcbC beta lactam genes in the marine fungus Kallichroma tethys. Applied and Environmental Microbiology, 69, p.1308–1314
Kohlmeyer, J. & Volkmann-Kohlmeyer, B., 1991. Illustrated key to the filamentous higher marine fungi. Botanica Marina 34, p.1-61.
Kohlmeyer J, Volkmann-Kohlmeyer B, 2003. Fungi from coral reefs: a commentary. Mycological Research,107, p. 386–387.
Magan, N., 2007, Fungi in Extreme Environments in The Mycota (Eds.) C.P. Kubicek &I.S. Druzhinina, Springer-Verlag Berlin Heidelberg 2007
Michaelis, K.C. et al, 1987. Population genetics and systematics of marine species of Dendryphiella. Mycologia, 79, p. 514–518.
Pugh, G. J. F. & Nicot, J., 1964. Studies of fungi in coastal soils. V. Dendryphiella salina (Sutherland) comb.nov. Transactions of the British Mycological Society 472, p. 263-267.
Spathas, D. H., 1978. A salt sensitive mutation on chromosome VI of Aspergillus nidulans. Aspergillus Newsletter 14, p.28.
Stanley, M. S., Hooley, P. & Clipson, N. J. W., 1995. A rapid method of heterologous gene cloning using cotransformation of lambda genomic DNA banks in Aspergillus nidulans. Fungal Genetics Newsletter 42, p.76-77.
Tresner, H. D. & Hayes, J. A., 1971. Sodium chloride tolerance of terrestrial fungi. Applied Microbiology 22, p.210-213.
Wethered, J. M., Metcalf, E. C. & Jennings, D. H., 1985. Carbohydrate metabolism in the fungus Dendryphiella salina. VIII. The contribution of polyols and ions to the mycelial solute potential in relation to the external osmoticum. New Phytologist 101, p.631-649.
Wilson, K. et al, 2005. Marine systems: moving into the genomics era. Marine Ecology 26, p. 3–16.
Yelton, M. M., Timberlake, W. E. & van den Hondel, C. A. M. J. J., 1985. A cosmid for selecting genes by complementation in Aspergillus nidulans : selection of the developmentally regulated yA locus. Proceedings of the National Academies of Science (U.S.A.) 82, p.834-838.