Summary
Viruses are microorganisms that can only survive and reproduce in the cells of other organisms and, as such, can infect a wide range of living entities, from bacteria to plants, animals, and humans. It is suggested that all the different viruses account for the quantity exceeding the number of stars in the universe (Wu, 2020). For this reason, the scientists developed various typologies that would help them to organize these microorganisms in groups based on common features and functions.
David Baltimore suggested one such classification, which sorted viruses based on how they produced messenger RNAs (Ryu, 2016). The first two groups include double-stranded DNA viruses and single-stranded DNA viruses. The next three groups encompass double-stranded RNA viruses, positive-sense single-stranded RNA viruses, and negative-sense single-stranded RNA viruses. Finally, single-stranded RNA viruses with a DNA intermediate and double-stranded DNA viruses with an RNA intermediate form the last two groups.
According to the other classification, viruses are grouped by the kind of cells that they infect. In this regard, bacteriophages are known as viral organisms that attach solely to bacteria. These viruses have received increased attention from the academic community since understanding the bacteriophage functioning can lead to their usage as human diseases treatment as an alternative to antibiotics. Considering that, all the current and future specialists in the spheres of biology and immunology need to have knowledge in how to examine these microorganisms in the laboratories.
The first step in this process is learning how to determine bacteriophage titer or phage concentration. To achieve that, the bacterial cells should be mixed with bacteriophage in soft agar in order to examine how the former would create colonies and the latter would form the plaque by destroying bacterial cells. Then, the plaque-forming units are determined based on the received plaque assay. Therefore, the aim of this report is to present the results of the experiment that followed the abovementioned procedure.
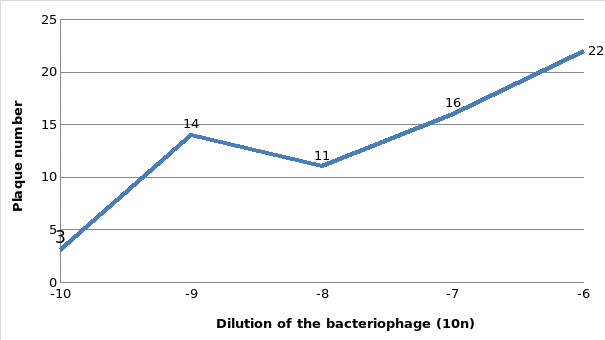
Results
Table 1: Dilution of the bacteriophage and plaque number
Reference List
Breijyeh, Z., Jubeh, B. and Karaman, R. (2020) ‘Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules, 25(6), p.1340.
Brzozowska, E. et al. (2018) ‘Interactions of bacteriophage T4 adhesin with selected lipopolysaccharides studied using atomic force microscopy, Scientific Reports, 8, pp. 1-7.
Durmuş, S. and Ülgen, K. (2017) ‘Comparative interactomics for virus–human protein–protein interactions: DNA viruses versus RNA viruses’, FEBS Open Bio, 7(1), pp. 96-107.
Engelman, A. and Cherepanov, P. (2017) ‘Retroviral intasomes arising’, Current Opinion in Structural Biology, 47, pp. 23-29.
Ge, H. et al. (2020) ‘The” fighting wisdom and bravery” of tailed phage and host in the process of adsorption’, Microbiological Research, 230, p.126344.
Ragland, S. A., and Criss, A. K. (2017). ‘From bacterial killing to immune modulation: recent insights into the functions of lysozyme’. PLoS Pathogens, 13(9), p. e1006512.
Ryu, W. S. (2016). Molecular virology of human pathogenic viruses. Cambridge: Academic Press.
Steward, K. (2021) ‘Lytic vs. lysogenic – understanding bacteriophage life cycles’, Technology Networks, Web.
Vergalli, J. et al. (2020) ‘Porin-mediated small-molecule traffic across the outer membrane of Gram negative bacteria’, Nature Reviews Microbiology, 18(3), pp.164-176.
Wu, K. J. (2020) ‘There are more viruses than stars in the universe. Why do only some infect us?’, National Geographic, Web.
Yoo, J. H. (2018) ‘Review of disinfection and sterilization – back to the basics, Infection & Chemotherapy, 50(2), pp.101-109.