Abstract
E.Z.N.A commercial kit is one of the greatest innovations in molecular biology. It helps in the extraction and purification of nucleic, deoxyribonucleic, and ribonucleic acids. The extraction is of great importance to researchers who aim at utilising deoxyribonucleic acid (DNA) from a variety of sources. The kit can be used in extracting the two molecules from their origins. The sources include plant materials, animal tissue, and soil. In this paper, the researcher sought to investigate the effectiveness of using the kit for the purposes of optimising the extraction of DNA from marine soils. To this end, two variables were analysed. The first was the quality of the acid yielded, while the second was the quantity that was extracted. Disruption of the cells was needed to obtain the acid. Bead beating was the process used to achieve this. Gel electrophoresis was also used to check for DNA fragmentation following this disturbance process. The research proved that the kit improved the quality of the DNA produced. The quantity of the material within a given volume of soil sample also increased. The kit was found to effectively extract DNA from an 8 gram sample of soil. The effectiveness was not achieved when a sample of 4 grams was used.
Introduction
DNA Extraction
The process is also commonly referred to as DNA isolation. It is the combination of both physical and chemical processes to extract the material from its various sources (Artyukhin & Woo 2012). The processes are aimed at purifying DNA generated from a sample. Today, the extraction is a common procedure in forensic and molecular biology. The procedure is of great importance since the isolated genetic material can be used to conduct further investigations in these fields. DNA extraction protocols can be divided into two major steps. The first step involves the gentle lysing of the cell to make the DNA soluble (Eldor 2007). The second stage includes a series of chemical and enzymatic reactions aimed at removing contaminants that may be present in the sample.
E.Z.N.A Commercial Kit
As already indicated, E.Z.N.A range of products is used for the purposes of extracting nucleic acid. The products were generated as a result of continuous innovation aimed at simplifying the process of removing and purifying nucleic acids from samples (Artyukhin & Woo 2012). A classic example of the products is the E.Z.N.A commercial kit. There are two types of nucleic acids. They are DNA and RNA. The former is the most important of these materials. The successful isolation of DNA is of great importance to researchers around the globe. There are various advantages associated with the use of E.Z.N.A commercial kits to extract DNA. One of them is the fact that the machine involves the use of optimised buffers. The buffers help in the extraction of pure DNA (Artyukhin & Woo 2012). In addition, the kit ensures that there are no organic contents in the sample. Lack of these elements reduces the number of impurities present in the extracted DNA. Genetic material gathered through the use of the kit is of high quality. It can be used for a variety of scientific investigations.
Another advantage of this pack is its duration of use. According to Eldor (2007), the end user is assured of a long period of service. The machine can work without interruptions for a period of one year. However, it must be stored under special conditions to ensure long life. For instance, OB Protease Solution should be stored at temperatures of 2-8°C. All other components of the kit can be stored at room temperature, ranging from 22°C to 25°C. The buffer solutions should also be checked for precipitates before they can be used (Oledzka 2012). Precipitates would make the gadget inefficient. To address this problem, the impurities are re-dissolved by raising the temperature of the sample to 37°C.
Extraction of DNA from the Soil
Microorganisms are the most ubiquitous of all creatures in the universe. They are present in almost all environments. All that is needed is for the environment to have the right conditions to support their existence. Some of the microbes are important sources of molecules that are of great importance in industrial and medical applications (Eldor 2007). A number of these microbes are also important sources of key enzymes used in the degradation of substances. As a result, it is important to isolate and study some of these important microorganisms living in the environment. The greatest challenge in doing this lies in the fact that the microbes do not exist in isolation.
Soil is one of the major reservoirs of microorganisms, especially prokaryotes. It is considered to be one the most complex environments. Microbes are heterogeneous and are distributed widely inside micro-aggregates and macro-porosities. The micro organisms are known to bind strongly to the soil particles (Mirnejad 2011). As a result, the whole microbe cannot be accessed. To be able to harvest the important molecules, it is important for one to isolate them from the soil particles.
A number of processes are undertaken to facilitate the extraction of these key molecules. To begin with, the soil particles are dispersed (Mirnejad 2011). The process of dispersal is aimed at dislodging cells that had been trapped in the inner compartments of the soil. It is important to consider the minute size of the prokaryotes when carrying out the dispersal to avoid their destruction. Centrifugation is then done. The main objective is to separate the bacteria from soil and organic particles. The process reduces the number of contaminants present in the extracted sample (Mirnejad 2011). The microbes are then cultured on various solid substrates. The plates used for culturing contain various nutrients to support the growth of specific bacteria only. Isolation and identification of individuals of a specific group is then done to ensure that microbes isolated are of pure composition. DNA extracts can then be sourced from the microbes.
The process is, however, complex and time consuming. Researchers invest a lot of time and resources in it. They are bound to make many mistakes while conducting the procedures (Boer et al. 2010). A lot of contamination is also likely to occur since no measures have been put in place to prevent the combination of the extract with other substances. Furthermore, the process requires a lot of manpower, making it a very expensive undertaking. The method also gives rise to a very small amount of DNA. Such an occurrence is associated with the inefficiencies linked to the procedures used (Boer et al. 2010). As a result, the procedure is not efficient where commercial extraction of soil DNA is needed. An alternative method is called for.
The use of E.Z.N.A commercial kit solves these issues associated with the extraction of soil DNA (Zhou 2011). To begin with, the process is done at a faster rate compared to the earlier method. Such levels of production can effectively meet market needs. Large quantities of DNA can be extracted at a go. The quality of the acid obtained is also high since little contaminants are present. The kit simplifies the work needed to achieve this. Less manpower is required for the extraction process (Boer et al. 2010). At the end of the day, the procedure is less costly to the researcher. The use of the kit also reduces the number of risks that a scientist is exposed to when handling reagents manually.
Materials and Methods
Materials
Soil Samples
- Marine soil samples with a moisture content of 54%
Equipment and Reagents
- Micro centrifuge capable of at least 14,000 x g.
- Nuclease- free 1.5ml microfuge tubes.
- Nuclease free 2ml microfuge tubes.
- Water bath preset to 65°C.
- Isopropanol.
- 100% ethanol.
- Vortexer.
- Incubator capable of 70°C.
- Centrifuge with a capability to hold 15ml centrifuge tubes.
- DNA solution isolated from the first procedure.
- Nanodrop ND-1000 spectrophotometer.
- Personal computer with Nanodrop 3.0 software.
- Purified water.
- DNA marker.
- Ice.
Methods
Prior to the extraction process, SPW wash buffer was prepared. The preparation was carried out by diluting SPW with 100% ethanol. The buffer was stored under room temperature. The heat elution buffer was set to 70°C. A bucket containing ice was prepared and the microfuge set at 4°C. 500 mg glass beads were added to a 15ml centrifuge tube. A starting sample of 0.4g of marine soil was added to the centrifuge tube containing the glass beads. 1ml of SLX-Mlus buffer was added and the contents vortexed using a mixer mill for 4 minutes. The aim of this process was to lyse the samples. The glass pebbles lyse the cells by breaking them open, releasing their intracellular components (Zhou 2011). Vortexing agitates the glass pellets to enable them lyse the cells.
100ul DS buffer was added to the lysed materials and mixed thoroughly using a vortex. The contents were incubated for 10 minutes at temperatures of 7°C. Later on, the contents were centrifuged at 3000 rotations per minute for three minutes. 800ul of the supernatant was placed in a 2ml microfuge tube, where 270ul P2 buffer was added. The contents were mixed thoroughly. The contents were incubated for five minutes. After this, they were then centrifuged at 14000 rotations per minute at 4°C for five minutes. The aim was to remove some of the proteins and contaminants. 70% isopropanol was then added. Mixing was then done by inverting the tube for 25 minutes. Isopropanol helped precipitate the DNA as it is not soluble in it. The DNA formed pellets by aggregating together after application of isopropanol. The contents were then centrifuged at 14000 revolutions per minute at 4°C for 10 minutes. The supernatant was drawn from the tube slowly to avoid upsetting the DNA pellet.
The tube was inverted on a filter paper to drain the fluid. 50ul elution buffer was added to the tube and the contents mixed thoroughly using a vortex. Incubation followed for 15 minutes. The aim was to dissolve the DNA granules. 100ul HTR reagent was added to the contents and left to rest for 2 minutes to remove inhibitors. Centrifugation at 14000xg followed for 2 minutes, after which the cleared supernatant was transferred to a new 2ml centrifuge tube. An equal volume of Xp1 buffer was added to the contents and mixed thoroughly. The mixture was transferred to HiBind DNA mini column after its insertion into a 2ml collection tube. Centrifugation at 14000xg for a minute followed, after which the filtrate in the collection tube was discarded. 300ul XP1 buffer was added into the solution. Centrifugation followed for one minute at 10000xg. The mini pilaster was relocated to a new cylinder. Here, 700ul SPW wash buffer was added. The procedure preceded centrifugation. The filtrate was discarded and the washing procedure repeated.
The empty Hibind DNA mini column was taken through a special treatment procedure. It was centrifuged at 14000xg for 120 seconds. The procedure was carried out at room temperature. The step was aimed at removing any residual ethanol that may be present in it. The pilaster was moved to a clean 1.5ml micro-centrifuge tube and 50ul elution buffer, which had a temperature of 7°C, was added into the HiBind membrane. Elution solution removes the DNA from the walls of the column and enables its collection at the end of the column. The contents were incubated for 12 minutes at 7°C. The incubation was followed by centrifugation at 14000xg for a minute. After this, the addition of elution solution and centrifugation was repeated. The eluted DNA was now ready for the next process of gel electrophoresis and spectrometer analysis. The procedure was repeated using samples of 0.4g, 0.42g, 0.41g, 0.8g, 0.88g, 0.86g, and 0.82g using 100ul elution solution
8% gel red containing 1ug/ml ethidium bromide, which acts as a stain, was the basis of analysing the extracted DNA. A number of materials and reagents were utilised in this procedure. They included an electrophoresis chamber, gel casting trays, and sample combs. Others were buffer (tris-acetate-EDTA), loading buffer, and a transilluminator. The procedure involved mixing the solution containing DNA with the loading buffer. The mixture was then placed into the sample wells. The lid of the electrophoresis chamber was applied and the power switched on.
The first procedure was to launch the software and lower pedestal with a soft wipe on the computer. The objective of this was to enable proper reporting. A blank sample was pipetted into the lower pedestal while the sampling arm was open. The sample chamber was closed by lowering the swing arm. On the screen, a click on the blank page launched analysis of the sample (Slater 2009). To reduce the probability of unreliable data, the process was repeated two times. On the cycle, a sample containing the extracted DNA was placed on the lower pedestal that was 1ul and the chamber closed.
After the sample was measured and the data printed, a procedure was carried out to determine the concentration. It was analysed through the use of a spectrophotometer to gauge its absorbance at 260nm (A260). In addition, A280 and A230 were measured. Ratios of A260 to A230 and those of A260 to A280 were used to measure the quality of DNA. The quality of this genetic material was measured by adjusting the A260 by absorbance at 320nm. The product of this was multiplied by the dilution factor. It was done through the use of A260 of 1.0=50ug/ml pure dsDNA.
Results
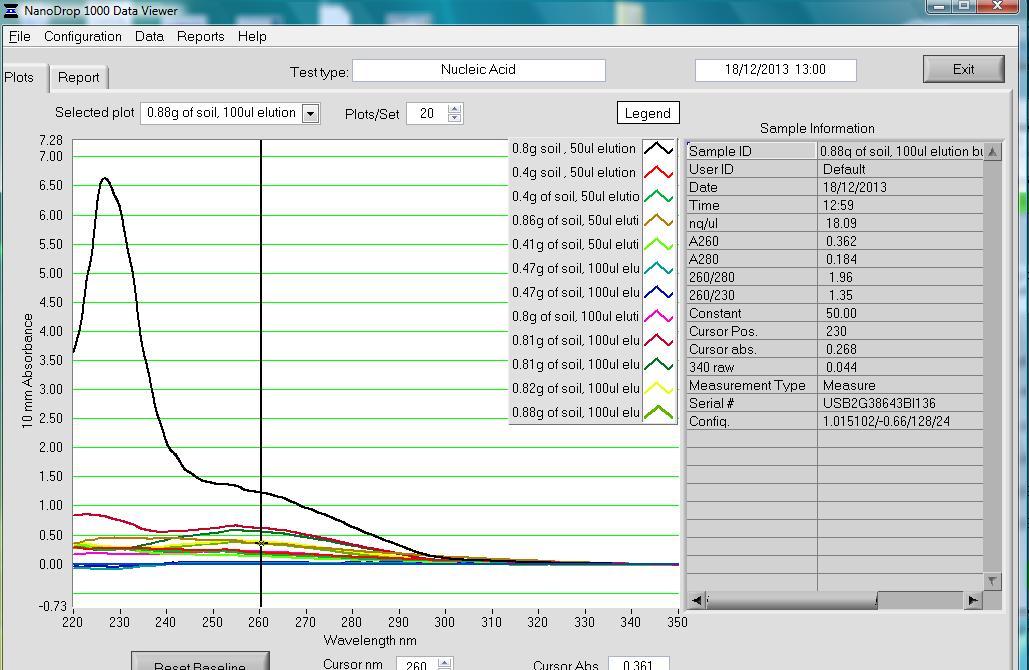
Results: This is first set of extraction using 0.47g and 0.42g soil volume and ≠0.5g of glass beads.
In this extraction, the researcher added 100ul elution buffer in one go (refer to step 37). However, only 50ul of the buffer was needed. For soil samples weighing 0.47g and 0.42g, there were no differences in the concentration of the extracts. However, the 260/280 and 260/230 absorption ratios for the 0.47g soil samples were higher.
The researcher then decided to repeat the experiment using 50ul elution buffer with the same volumes.
It was found that the results for the 0.4g soil volume were not very pure. Impurities were noted regardless of whether the 50 or 100ul elution buffer was used. As a result, the researcher repeated extractions using 0.8g volumes and 50ul elution buffer to see if there were any differences.
Purity had improved and the concentration of DNA was higher using 0.8g volume of soil.
The researcher decided to do 4 extractions using 0.8g volume of soil. The aim of this was to analyse the differenced. In these procedures, the 100ul elution buffer was used.
A 260/280 absorption of between 1.7 and 2.0 is good. As such, the 0.8 samples showed that they contained a high qualities of the DNA extracted. However, concentration was not consistent.
The 29.8ng/ul concentration of DNA seemed quite high compared to the 9.4ng/ul, even though the volumes were similar. It was assumed that perhaps the sample used had other DNA from plant roots mixed with the soil. It was concluded that the 0.8g was the suitable volume to use compared to the 0.4g. It was also inferred that the 100ul elution buffer was better compared to the 50ul one. Based on the peaks from the chromatograms, it was decided that it may be guanidine contamination. To resolve this problem, the researcher decided to add an extra wash step.
Steps 31-33 repeated.
Here are the results:
The results above include those made in the extra wash step. It appears that they are negative. What this means is that the DNA was possible washed out. The gel electrophoresis showed the same. There are no bands visible on the gel red.
Discussion
The E.Z.N.A commercial kit is a very important tool to molecular scientists. The machine is crucial in the isolation of DNA that is of high quality and in large quantities. Both quality and quantity of extract are important aspects of research (Artyukhin & Woo 2012). The large quantities of DNA extracted through the use of the kit shows the level of efficiency associated with it. As a result, the amount extracted can be used in a variety of studies. Furthermore, the products can be stored or cultured for further studies. The kit also ensures that the long process of extraction is economical. To this end, the amount of DNA extracted is commercially viable.
The quality of DNA is another element that must be considered. The final product must be free of any contaminants for the right results to be obtained when conducting experiments. Consequently, washing is considered to be an important step when using the kit. It ensures that all potential contaminants are removed (Artyukhin & Woo 2012). The slightest mistake in the purification process can result in huge variations in research findings. The E.Z.N.A commercial kit is capable of purifying the DNA extracted from a variety of sources, including marine soils (Nannipieri 2006). For this reason, the kit has been widely recommended for commercial extraction of the material.
In this study, the E.Z.N.A commercial kit was seen to better extract DNA from a 0.8g sample as compared to from 0.4g. Efficiency was observed in terms of increased quantity and quality of the material produced. When 0.4g of marine soil was used, the results obtained indicated inefficiencies, even after both 50 and 100ul elution buffers were used. The reason behind this is that 0.8g of soil has a considerably higher composition of DNA compared to a sample of 0.4g (Eldor 2007). As a result, the reagents utilised were put to maximum use since there was enough cell material to lyse. However, extraction of DNA from a 0.8g is advantageous to the researcher in that fewer repeat processes would be done compared to a smaller sample. In this case, a single extraction using 0.8g of soil would save time. Two samples of 0.4g are needed to generate the same matter as one sample of 0.8g.
Absorbance measurements are used to determine molecules that absorb matter at specific wavelengths. The molecules should first be purified to ensure that the right results are obtained (Wang & Son 2013). In this case, absorbance can be used to analyse the purity of these molecules.
The 260/280 absorbance ratio is very important. It is used to evaluate the purity of the two genetic materials. A value of 1.8 is expected for DNA, while RNA is set at 2.0. A value that is lower than this may be an indication of the presence of a contaminant. In this case, the source of contamination may be a phenol or a protein. It may also be any other substance that absorbs at a wavelength of approximately 280nm (Zhou 2011).
Absorbance at 230 nm is widely recognised as the wavelength at which other contaminants take in substances. As a result, the absorbance ratio of 260/230 is used when analysing the purity of nucleic acids. The value of pure acid is usually higher than that observed using 260/280. The values obtained from this ratio vary between 2.0 and 2.2 (Zhou 2011). Lower values may be as a result of contaminants that absorb at 230nm and below. They include glycogen, phenol, carbohydrates, and guanidine.
Guanidine is a colourless solid that is soluble in polar solvents. It is a strong base. The substance is obtained from natural sources, usually as a result of the oxidation of guanine. The oxidant is one of the nitrogen bases that make up the nucleic acids (Wang & Son 2013). Both DNA and RNA contain guanine. In the case of this experiment, the substance may result from the fragmentation of nucleic acids as they are being extracted.
From the results obtained, it is obvious that the use of 0.4g of soil showed evidence of extraction. However, the amount of DNA generated was low. Absorption ratios were also quite low. The purity of the product was likewise low, something that affected its quality (Eldor 2007). The poor quality of the DNA can be attributed to guanidine contamination. The use of 0.8g of soil, however, produced better results than the ones obtained in 0.4g. The quality of the matter extracted was also high.
The experiment was repeated five times using the 0.8g of soil. However, the results obtained showed a lot of inconsistence in terms of the quantity of DNA that was extracted. For instance, one experiment showed extraction levels that were as high as 29.8ng/ul, while another showed low levels of 9.4ng/ul. The findings were made despite the fact that similar conditions had been set for the two experiments. The amounts of reagents used were also similar.
The variations in the amount of DNA generated can be attributed to the fact that the different portions of the soil samples used may have varying compositions of organisms. The soil is known to harbour a number of macro and microorganisms. The former have a higher composition of DNA compared to the latter (Boer et al. 2010). In addition, the soil may contain other organic materials. They include such elements as roots and other decomposing plant matter. The additional materials also contain DNA. Extraction of genetic matter from such materials will give false results. The DNA produced by such soils, though of high quantity, may be of little use to the researcher (Verollet 2008). However, the purity of the acid extracted from the 8.0g soil volumes was high despite the inconsistencies in the quantities observed.
The low quality of the DNA extracted in the experiment that used 0.4g can be put down to a number of factors. One of them is the fact that only a single wash was done using the SPW buffer. Absorption ratios were also considerably low (Zhou 2011). The observation can be as a result of using the 50ul instead of 100ul elution buffer. For the case of the 0.8g soil sample, an extra washing step was applied. The special treatment possible explains the high quality of DNA that was yielded. The small quantity of the DNA that was extracted following the use of the 4.0g soil can be associated with the use of the 50ul elution buffer (Zhou 2011). The work of this reagent is to remove DNA from the walls of the column. The fact that 100ul elution buffer was used in the case of the 8g soil sample means that there was a better removal of the DNA present on the walls, which was not the case with the 4.0g slide. As a result, a larger quantity was likely to be collected from the former sample in comparison to the latter.
Gel electrophoresis is a technique used for the purposes of analysing DNA. The process is used to check for fragmentation within the DNA strands. No fragmentation was observed when the samples used in this experiment were taken through this procedure (Slater 2009). For this reason, an extra wash step was conducted. However, such washing can expose the DNA to fragmentation. The additional cleaning step was aimed at removing contaminants from the genetic material collected. Contaminants would include such substances as guanine and other cell contents. Traces of soil may also be present. SPW wash buffer was the reagent used to clean the DNA samples obtained following extraction (Zhou 2011).
When the experiment was repeated, it was found that the chromatograms had shifted their peaks. They were not similar to those of pure DNA. The aspect can be attributed to contamination (Slater 2009). Guanidine staining is the most possible reason for this finding. As a result of bead beating, the DNA molecules were fragmented, leading to further contamination. The development is what brought about the changes in peaks
In an experiment that included an extra wash, the results showed that no DNA was present in the sample. The most possible reason behind this observation would be that the SPW wash buffer destroyed the structure of the DNA, leading to fragmentation (Zhou 2011). The occurrence is due to prolonged exposure to the reagent. Once the DNA molecules are destroyed, their sizes are reduced. In addition, their content is likely to dissolve in the wash buffer (Wang & Son 2013). When the tube is inverted over the filter paper, the DNA material is also lost in the process since it is able to percolate within the tiny pores of the material.
Bead beating is a mechanical method of disrupting cell structure. The technique, which is also known as milling, utilises particles made of glass, steel, ceramics, or tungsten carbide (Verollet 2008). The type of pebbles used depends on the type of organism whose cells are to be disrupted. For bacteria, the beads are usually made of glass and are 0.1 to 0.6mm in diameter. For yeast and other unicellular animal cells, they are 0.5mm in diameter. Plant and animal tissues are disrupted using either tungsten or stainless steel. The beads are usually 3-7mm in diameter. The basic procedure for disrupting the cells involves placing the sample, mixed with the beads, in a test tube or a bead beater machine. During agitation, the pebbles collide with the cellular material, tearing them apart (Wang & Son 2013).
The level of mechanical shear is moderate. However, in other cases, the disruption is extreme. The most efficient bead beaters are able to generate forces that make it possible to perform three different grinding processes at the same time. The three processes include cascade impaction, which resembles a hammer-like process of working on the sample (Verollet 2008). The second is shearing as a result of the sharp edges of the beads colliding with the solids in the sample. The granules act like trajectories and are able to puncture the cells, especially when the process is carried out in a medium (Boer et al. 2010). The last process is the creation of a vortex-like movement generated by the agitation of the materials in the machine. In small labs, equal amounts of sample and pellets are placed in a test tube and mixed vigorously using a common laboratory vortex (Verollet 2008). When the procedure is carried out using this machine, the process is about 3 to 10 times lower compared to the other apparatus. The advantage of the system is that it is inexpensive and can yield a breakage rate of about 50%.
In more advanced laboratories, bead beating is carried out using automated machines. The machines can agitate the material at around 2000 oscillations per minute (Verollet 2008). In most cases, the machines are connected to strong electric motors. The machine completes disruption in less than three minutes (Boer et al. 2010). In contrast to other mechanical methods, bead beaters are able to work on small portions of a sample.
Bead beating has a number of major disadvantages. One of them includes heating the components. Agitation of the pellets leads to an increase in the amount of heat in the media due to friction. There are various ways to circumnavigate this problem. First, after the disruption, the sample can be cooled by fusing it with ice to prevent damage to proteins and enzymes (Verollet 2008). Second, the sample may be cooled by alternating the bead beating and cooling processes. In addition, an air coolant may be circulated in the bead beater as the process takes place. Another method of increasing the efficiency of these machines is through the addition of solvents prior to beating (Boer et al. 2010). The solvents dissolve the cell wall and the cell membrane, making it easy to disrupt any existing cell structures.
Bead beating is the process that was applied in the disruption of cells in the E.Z.N.A commercial kit (Boer et al. 2010). However, care should be taken when carrying out the procedure. Control measures should be put in place to ensure that the optimum results are obtained from the experiment. To begin with, oscillations used should be about 2000 revolutions per minute. Low speeds may reduce the efficiency of the process, while high revolutions may lead to mild injury of the cell. The latter would result in the destruction of the DNA strands (Verollet 2008). Higher oscillations would also lead to excessive heat production, which would lead to the fragmentation of DNA. Enzymes would also be denatured in the process. Fragmentation would reduce the yield of the experiment. Guanidine contamination would also occur in the process. Cooling is important to reduce the effects of heat on the DNA samples extracted.
260/230 is one of the best absorption ratios. It enables the researcher to account for contamination resulting from guanidine. The ratio improved the results obtained from the extraction process (Nannipieri 2006). Based on the findings made in this experiment, it was determined that the values obtained from the 260/230 ratio were lower than those obtained from the 260/280 part. However, the results for pure DNA should reflect a higher value using the 260/230 ratio compared to the 260/230 proportion. For this reason, the quality of the DNA extracted was very low.
One of the samples showed absorbance ratios that were higher than the 1.8-2.0. Such values are expected when the 260/280 ratio is used. The sample contained 0.88g of soil. However, the value obtained from the 0.82g sample fell within the expected range. On their part, the 0.80g and 0.81g samples had lower values than those of 0.82g.
Conclusion
DNA is one of the most important biological molecules (Nannipieri 2006). The molecule is used in a number of studies, especially those associated with molecular biology. Extraction of the molecule is regarded by researchers and other scholars as a difficult undertaking. The development of the E.Z.N.A commercial kit was meant to address these issues. The machine was meant to help researchers around the globe to obtain this key molecule with relative ease. With the help of the kit, DNA can be extracted from a wide range of sources.
References
Artyukhin, A & Woo, Y 2012, ‘DNA extraction method with improved efficiency and specificity using DNA methyltransferase and “click” chemistry’, Analytical Biochemistry, vol. 425. no. 2, pp. 169-174.
Boer, R, Peters, R, Gierveld, S, Schuurman, T, Kooistra-Smid, M, & Savelkoul, P 2010, ‘Improved detection of microbial DNA after bead-beating before DNA isolation’, Journal of Microbiological Methods, vol. 80. no. 2, pp. 209-211.
Eldor, P 2007, Soil microbiology, ecology and biochemistry, 3rd edn, Academic Press, London.
Mirnejad, R 2011, ‘Rapid DNA extraction of bacterial genome using laundry detergents and assessment of the efficiency of DNA in downstream processes using polymerase chain reaction’, African Journal of Biotechnology, vol. 11. no. 1, pp. 209-211.
Nannipieri, P 2006, Nucleic acids and proteins in soil, Springer, Berlin.
Oledzka, I 2012, ‘Comparative evaluation of tissue protein separations applying one- dimensional gel electrophoresis and capillary gel electrophoresis’, The Open Proteomics Journal, vol. 5. no. 1, pp. 17-21.
Slater, G 2009, ‘DNA gel electrophoresis: the reptation model(s)’, Electrophoresis, vol. 30. no. 1, pp. 181-187.
Verollet, R 2008, ‘A major step towards efficient sample preparation with bead-beating’, Biotechniques, vol. 44. no. 6, pp. 832-833.
Wang, X & Son, A 2013, ‘Effects of pretreatment on the denaturation and fragmentation of genomic DNA for DNA hybridization’, Environmental Science: Processes & Impacts, vol. 15. no. 12, p. 2204.
Zhou, C 2011, DNA binding and DNA extraction: methods, applications, and limitations, Nova Science, Hauppauge.