Introduction
The laboratory tests allow for the identification of the difference between gram-positive and -negative bacteria in practice. While gram-negative bacteria lose crystal violet and stain red, gram-positive bacteria stain purple and keep crystal violet (General Microbiology Laboratory Manual, 2022). Gram staining can thus differentiate between the two types of bacteria.
Due to their thick cell walls, gram-negative bacteria are more immune to antibodies (General Microbiology Laboratory Manual, 2022). Recently, the importance of gram-positive bacteria in freshwater ecosystems and their presence in marine environments have been recognized (General Microbiology Laboratory Manual, 2022). However, their quantitative value and potential involvement in these environments, mainly coastal areas, still need to be determined. Gram-negative bacteria are almost solely found growing in water and diluted solutions because they are adapted to poorly osmolarity settings (General Microbiology Laboratory Manual, 2022). These distinctions are critical in the evaluation of the laboratory test results.
When oral microbes break down sugars to create acid, which demineralizes the teeth’s hard tissue, dental caries, both dentine and enamel, are formed. Bacterial acid that dissolves the tough dental structures causes cavities (Saheb et al., 2022). Acid is generated when microbes digest leftover food or sugar on the tooth. Bacteria that can produce acids through their chemical reactions are known as acidogenic bacteria (Saheb et al., 2022). Streptococcus epidermis is the primary acid-producing bacterial species in dental cavities.
While coliforms by themselves typically do not result in severe disease, they are simple to culture, and their presence is used to suggest the presence of other pathogenic organisms with fecal genesis (Renter et al., 2022). Coliforms can be found in soil, water, and on plants. The likelihood of contracting a disease transmitted by water is raised if coliform organisms are present in the water people consume.
Although total coliforms can originate from sources other than waste, a positive overall coliform sample should indicate contamination. A confirmed coliform test suggests potential pollution and a threat of illness caused by water (Renter et al., 2022). More tests for fecal coliforms or E. coli are typically mandatory after a positive test result for total coliforms.
Materials and Methods
The samples used for the experiment come from water, throat, and skin. A throat test was performed on me, and a skin test sample, mainly the test from the face (nasal skin), was taken from the lab partner. The results show that the bacteria on her face were Streptococcus epidermidis. The water for the sample used in this laboratory report comes from the Layette River, where the coliform bacteria were found.
The samples were used to determine the bacteria and their type based on the results of the tests. It is possible to decide what kind of microorganism was identified after performing such actions as isolation, subculturing, pure culture, gram stain, catalase test, and coagulase test. The length of incubation of coliform bacteria is 12-72 hours, and the temperature ranges from 35 to 37°C; the air is the primary incubator of the coliform bacteria (Renter et al., 2022)
Staphylococcus epidermidis can survive under a wide range of temperatures, but its optimal temperature range is 30 to 37°C. Its incubation period lasts from 4 to 10 days, which influences the process of the laboratory experiment (Peng et al., 2021). The following major tests were used during the laboratory experiment.
Gram Staining
A pure culture was obtained and distributed on the plate after the inoculation loop was disinfected; attention was required to avoid combining the extract. After placing the culture filtrate on the scale, it was heated for a few seconds with a lighter to mend it. The first stage involved applying the first stain, crystal violet, directly to the slide for about 30 seconds.
After the solution had been on the decline for 30 seconds, it was carefully rinsed with distilled water. Iodine solution was used for the mordant in the subsequent stage and was applied for 60 seconds. The third stain was an ethyl alcohol wash, which was applied and kept on the slide’s exterior for twelve seconds after saturation and another water rinse.
The alcohol cleanser was rinsed with water again. Last but not least, 60 seconds had passed since the safranin counterstain was administered. The slide was then thoroughly washed. Finally, 60 seconds had passed since the safranin counterstain had been distributed. To remove any remaining stains, a final washing with water was used.
After the experiment, the slide was examined under a bright field microscope at a magnification of 1000x to determine whether the color exhibited was reddish/pink to distinguish between bacteria. Compared to gram-positive bacteria, which would have appeared purple, they suggested they were gram-negative.
Urea
To correctly inject culture, the primary culture tube was shaken to suspend bacteria. A sample was collected and put inside the Urea test tube after the inoculation loop had been sterilized, and it was then incubated for 48 hours. Pink, which represents robust urease synthesis, would be a favorable outcome. The sample would not have contained urea hydrolysis if it had tested negative and had no pink hue.
Citrate
After a clean inoculation, a needle was used to collect a bacteria sample, and a BBL Simmons’ Citrate Agar Slant tube was inoculated using the slant and stab method. The test container was then kept at 37°C for 48 hours. The test aimed to determine whether bacteria could develop using citrate.
Nitrate
The Nitrate test required cultivating the bacterium in a culture test tube employing Durham tubes, nitrate broth, and 0.5% potassium nitrate. Bacteria were transferred from the culture using a sterile loop and gently tapped inside the test tube to ensure the sample was obtained correctly. A 48-hour incubation phase took place at about 37°C. The test tube was inspected after incubation to determine whether any specific growth had occurred. The gas bubbles during the Nitrate test would have indicated an acceptable result, demonstrating that an unidentified bacterium had converted nitrate to a gaseous end product.
Negative outcomes of the process would have been present if there had been no gas bubble formation, resulting in the organism’s nitrite production or other nongaseous products. The culture tube was mixed with about 0.5 mL of the nitrate diagnostic reagent to look for a potential color shift. If the red or pink color appeared, it would have been proof that the medium was negative and affirmed that the outcome was positive. To validate a negative test result, 5-10 drops of zinc powder would be added, and the tube would be gently swirled. The outcome will depend on the color change; the altered color indicates the nitrate presence.
FTM
The experiment found unidentified microorganisms and determined whether the bacteria needed air to develop. The unidentified organism’s aerotolerance would have eventually determined whether it was aerobic (requires oxygen to survive). Three tests were performed to determine the unknown oxygen capability or requirement for oxygen use. Three tests—MR-VP, Citrate slant, and Nitrate with Durham tube—were performed to confirm the oxygen requirements for the unknown bacteria. Different outcomes of these suggested procedures would support the conclusions of the undetermined material.
Methyl Red Test
A tube of BBL MR-VP broth was inoculated with an unidentified bacterium using a sterile loop for this Methyl Red test. It had undergone a variety of experiments and been incubated for 48 hours at a temperature of 37°C. After incubation, one-third of the bacterial colony was transferred into a different test tube. Then, there were added five droplets of methyl red indicator. Following the addition, observations were made to determine whether the bacteria could ferment the glucose and reduce the pH of the solution.
Voges-Proskauer Test
The sample containing methyl red was divided by about one-third and placed in a different test container. Then, while shaking the test tube to integrate aeration, fifteen drops of Barritt’s solution A and five drops of solution B were added to each culture. Within the test containers, reactions would happen instantly or within 20 minutes. Red would be associated with successful test findings. The test finally concentrated on the unidentified bacterium’s capacity to ferment glucose and produce acetyl methyl carbinol.
Results
Finding the differences between bacteria found in gram-positive and gram-negative cells requires several stages. Because bacteria vary chemically and physically, and in terms of how they respond to the staining process, differential staining is necessary to identify the type of bacteria. Figures 1 and 2 depict an unknown organism at various magnifications. Following Gram staining, it was discovered that the unidentified bacterial cells displayed a pinkish-red color and a rod-like form, emphasizing each distinct cell. Table 1 reflects the laboratory tests required for identifying the bacteria.
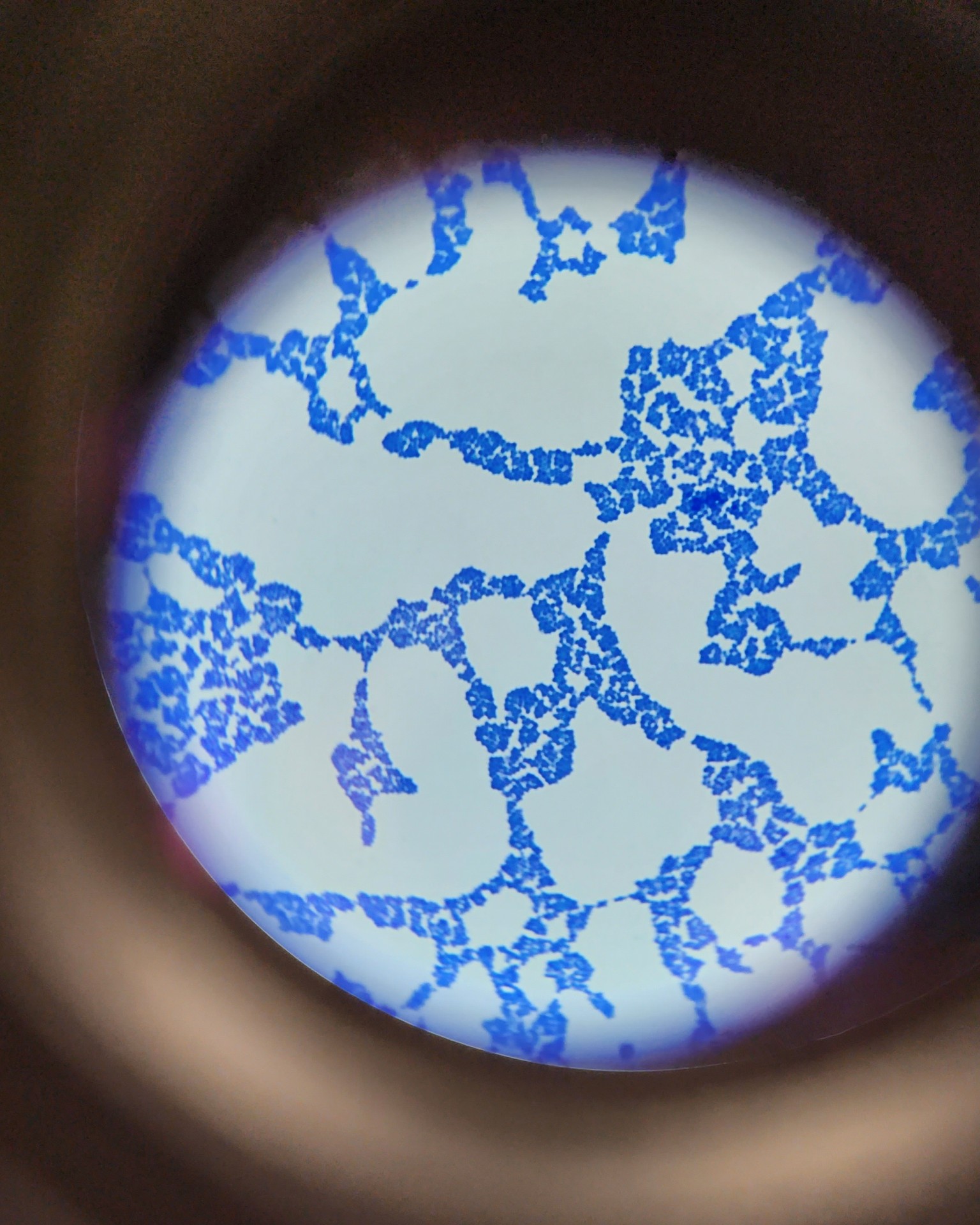
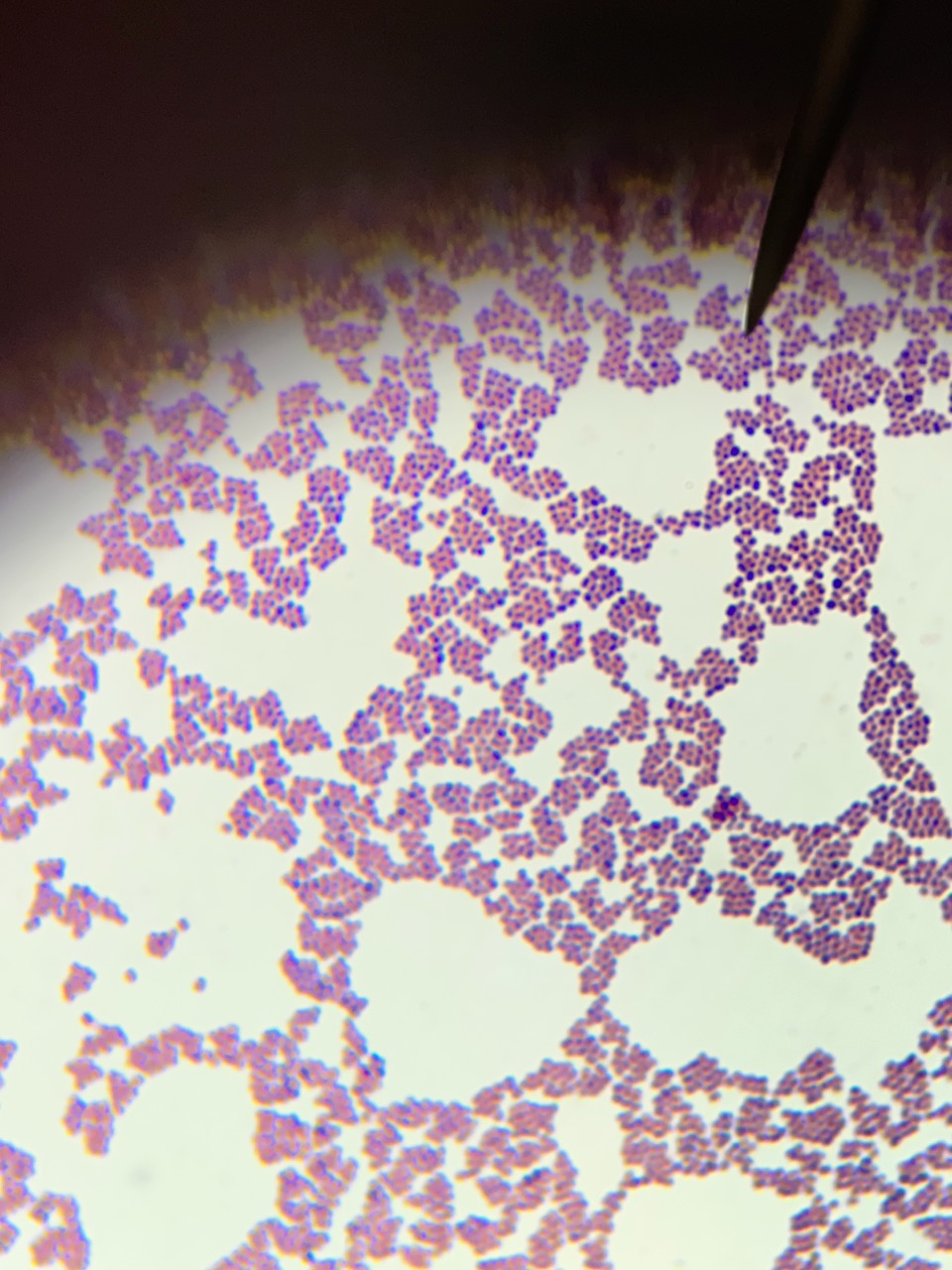
The previous images illustrate the conventional biochemical methods for identifying unknown bacteria. After Gram staining, there was evidence of a gram-negative bacterium being present. Unknown microorganisms showed rod-shaped under magnification after Gram staining (Figures 1 and 2). Positive findings from the citrate test indicated citrate usage by the appearance of a blue color (General Microbiology Laboratory Manual, 2022).
Table 1: Tests for Identifying Bacteria
Gram Stain
Urea Hydrolysis
Simmon’s Citrate
Nitrate Reduction
Methyl Red Test
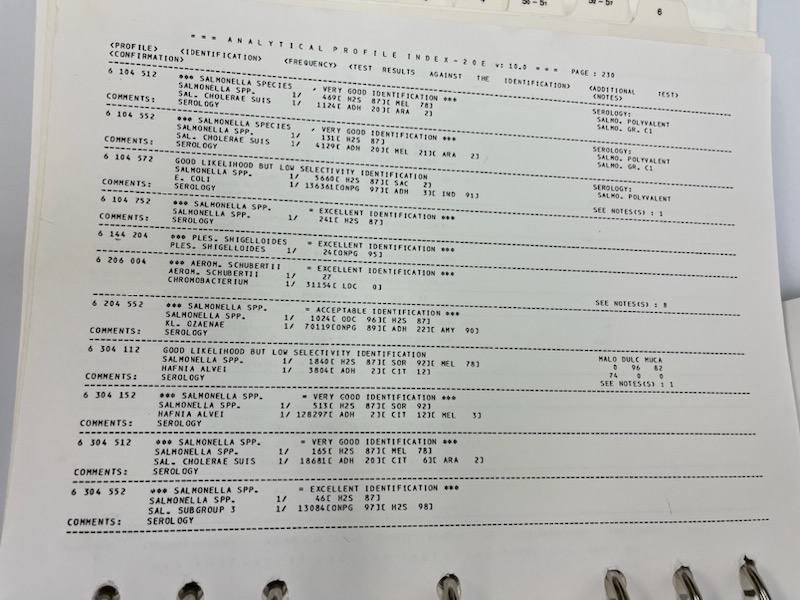
Poor urea hydrolysis, nitrate reduction, FTM, and methyl red results revealed an unknown organism (General Microbiology Laboratory Manual, 2022). After completing all processes, the results obtained using the Manual of Determinative Bacteriology showed that the gram-negative coliform bacteria and an unknown bacteria, most likely Streptococcus epidermidis, were present.
Discussion
The microorganism is the cell that scientists can see only using the microscope, which makes laboratory research critical. Using the genus-species formula when identifying an organism is essential. In a binomial terminology system, the species is the particular name, while the genus is the generic name (Peng et al., 2021). It facilitates understanding the bacteria and their classification into gram-positive and gram-negative.
Microbiology benefits greatly from identifying bacteria because it allows for the early detection of dangerous types and the control of their spread. Due to technological advancements, numerous methods are available when experimenting with an unidentified bacterium. The presence of a particular bacterium is investigated in lab environments using conventional molecular and biochemical techniques. These methods continue to be used in contemporary laboratory environments and frequently offer scientists conclusive information concerning unidentified organisms. These approaches can also avoid erroneous results, so experiments must be repeated multiple times to justify the objectivity of their conclusions.
Molecular methods can deliver multiple organisms in the context of a single study, making them one of the different tests that can be performed on an unknown bacterium. The molecular approach’s sufficiency involves sufficient selectivity and sensitivity and can quickly outperform culture-based methods in general. Chemicals distinctive to a given species can be found using a biochemical test. The test is a time-tested and cost-effective way to evaluate an organism. The biochemical method, which typically incorporates test tubes and plates to analyze bacteria, is helpful when identifying types of microbes while using a manual to find critical distinctions.
After studying the theoretical material for the discussed topic and conducting laboratory experiments, I concluded that laboratory experiments are crucial in microbiological investigation. The identification of unknown bacteria is frequently used in the laboratory for many reasons. Bacterial identification is most commonly used during epidemics, especially in environments where nutrition is distributed.
To stop contamination and ensure appropriate food distribution, the type of bacteria causing the outbreak must be identified before it can be prevented. The unknown bacteria were placed in the lab using well-known traditional biochemical methods, including urea, citrate utilization, FTM, methylene red (MR) indication, nitrate reduction, hydrolysis, and the Voges-Proskauer (VP) test. The test findings showed positive MR, citrate, negative urea, nitrate, and aerobic bacteria.
Conclusion
The results indicated that the gram-positive bacterium that is the pathogen of various infections was Streptococcus epidermidis, while the gram-negative one was the coliform bacteria. Therefore, the microorganisms have been adequately identified and tested. It is difficult to state that something needed to be fixed during the experiments because the team followed the steps of conducting the experiments attentively.
References
General microbiology laboratory manual. (2022). Biol317. [PDF file].
Peng, X., Zhu, Q., Liu, J., Zeng, M., Qiu, Y., Zhu, C., Cheng, Y., Zhou, Y., Xu, Y., Chen, M., Wen, Z., Chen, Y., Li, R., Tong, J., Shan, Q., Lin, D., Wu, S., Zhuo, Z., Wang, C., Zhao, S., … Collaborative Working Group of the Pediatric Subgroup of the China Society of Infectious Diseases. (2021). Prevalence and antimicrobial resistance patterns of bacteria isolated from cerebrospinal fluid among children with bacterial meningitis in China from 2016 to 2018: a multicenter retrospective study. Antimicrobial Resistance and Infection Control, 10(1), 24. Web.
Renter, D. G., Dodd, C. C., Noll, L. W., Nagaraja, T. G., & Ives, S. E. (2022). Coliform and Escherichia coli Contamination on External and Internal Surfaces of Beef Carcasses with and without Tissue Adhesion Excision. Journal of Food Protection, 85(4), 701–705. Web.
Saheb, K., S., Proctor, D. M., Deming, C., Saary, P., Hölzer, M., NISC Comparative Sequencing Program, Taylor, M. E., Kong, H. H., Segre, J. A., Almeida, A., & Finn, R. D. (2022). Integrating cultivation and metagenomics for a multi-kingdom view of skin microbiome diversity and functions. Nature Microbiology, 7(1), 169–179. Web.