Summary
In the last 15 years, there has existed a cosmopolitan increased correlation between vancomycin-resistant enterococci (VRE) and severe clinical infection “(1, 2), forcing hospitals to set up counteractive measures to control the pandemic. To aid in countering the epidemic, they uncovered gut-colonized patients who were non – infected and would serve as the epicenter in the spreading of VRE. Furthermore, employing the use of culture-based techniques will go a long way in alleviating this challenge. The experimentation scientifically evaluated the performance characteristics of two culture-based chromogenic media that are cost-effective for VRE surveillance” (3, 4). The outcome of the results was based on Sensitivity, Stability and Precision as the three parameters that facilitated the analysis of Enterococcus species into two sets of media for selectivity determination. For control monitoring, positive control strains from VRE reference (n=4) from two different sources were also cultured. A dilution of two Enterococcus spp, Enterococcus faecium n=7; and Enterococcus faecalis ;n=2) reference and streaking on unadulterated and assorted cultures of both Media, followed by a 48-hour incubation.
To clearly outline this issue, Vancomycin-resistant enterococci organisms were analyzed and comparisons evaluated using two Chromogenic (CHM-Gn) media. The outcome of the experiment was impressive where the two media, CHM-Gn “A” and CHM-G “B” produced exemplary results after they were enriched overnight and incubated for 48-hours. Both media had 98% (53/54) sensitivity and an average of 95.5% (196/205) specificity. Biochemical testing was used to further identify species whose color was difficult to pick especially on CHM-Gn “A”. A light microscope was also used to observe the easily unidentifiable species to the naked eye.
Media “A” and “B” produced exemplary results after they were enriched overnight and incubated for 48-hours. Both media had a 98% (53/54) sensitivity, 95.5% (196/205) specificity. Both species Enterococcus solitarius and Enterococcus hirae were unable to grow on both media. Enterococcus gallinarum was able to grow as tiny bluish colonies on CHR Upon dilution and 48-hours incubation. Both media revealed a typical colony appearance as described by the manufacturers (Table A). The presence of foul bacteria on the plate resulted in low sensitivity.
After 24 hours of incubation unexpectedly, with seven color shades at colony edges for VREfaecium and six for VREfaecalis. Two similar colors were observed in the center of VREfaecium and VREfaecalis colonies, thereby complicating definite VRE species identification. Generally, after 24hrs cultivation, VREfaecalis presented a stable green color while VREfaecium had a different mixture of colors. VREfaecium colonies were seen as red most of the time. Colonies on “A” expressed superior growth in comparison to “B” colonies leading to untimely uncovering and identification of VREfaecium and VREfaecalis in 24hrs. Xylose fermentation test confirmed E. gallinarum species growth on media “A” and “B”. Yeast also cultivated well as white (on “B”) and pale green (on “A”) media as that had led to misleading colony colors.
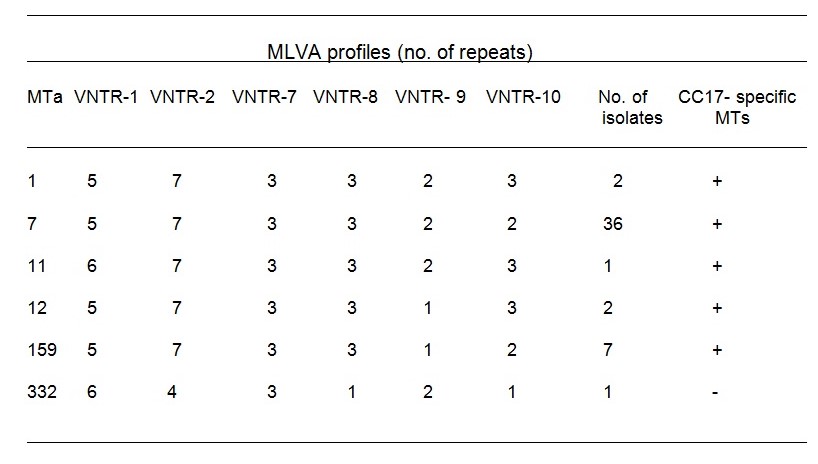
Comparative analysis revealed unacceptable results after foul bacteria were cultured, hence one of the VRE spp undetected from the two media. This was as a result of plating saline diluted stool samples on media “A” and “B” Comparable screening of over 200 stool specimens on media “A” and “B” was analyzed, resulting in a colonization index of 0.21(21%) for both media (Table A). The MLVA (Multiple Locus Variable Analysis) was used to determine VREfaecium’s genetic relations among its isolates(Over 40 isolates), where a summation of six unlike MLVA variety was arrived at (5). These varieties were expressed in both media.
The experimentation clearly revealed that both CHM-Gn media don’t absolutely fulfill prospects considering VRE exposure, particularly when inoculating stool samples straight or following pre-dilution. Irresistible cultivation of diverse bacteria organisms and yeasts on media “A” and “B” hinder agar assessment while culturing stools samples directly. Stool samples are therefore unsuitable for straight inoculation onto both media. “Overnight stool pre-inoculation to facilitate enrichment is paramount prior to plating samples onto both media (3). Projected major reactions support the key classification of doubtful isolates, hence, therefore, greatly recommended in shunning pseudo positive outcomes and verification of vancomycin resistance.
Analysis of colony color “B” revealed colony color stability during the 24 and 48 hrs growth periods. This is however unexpected according to CHM-Gn “A” manufacturers. On “A” medium, the appearance of different color shades of VRE species further complicates the analysis. Regrettably, instituting the phase of enrichment slowed VRE identification. Fortunately, using either media presents a VRE cost-cutting screening technique for stool analysis, hence safety measures decreasing detrimental infection prevalence.
Critical Review
Abstract
Vancomycin-resistant enterococci organisms were analyzed and comparisons were evaluated using two Chromogenic (CHM-Gn) media. The outcome of the experiment was impressive where the two media, CHM-Gn “A” and CHM-G “B” produced exemplary results after they were enriched overnight and incubated for 48-hours. Both media had 98% (53/54) sensitivity and an average of 95.5% (196/205) specificity. Biochemical testing was used to further identify species whose color was difficult to pick especially on CHM-Gn “A”. A light microscope was also used to observe the easily unidentifiable species to the naked eye.
Introduction
“In the last 15 years, there has existed a cosmopolitan increased correlation between vancomycin-resistant enterococci (VRE) and severe clinical infection “(1, 2), forcing hospitals to set up counteractive measures to control the pandemic. To aid in countering the epidemic, they uncovered gut-colonized patients who were non – infected and would serve as the epicenter in the spreading of VRE. Furthermore, employing the use of culture-based techniques will go a long way in alleviating this challenge.
Materials and Methods
“The experimentation scientifically evaluated the performance characteristics of two culture-based chromogenic media that are cost-effective for VRE surveillance” (3, 4). The outcome of the results was based on three parameters that facilitated the analysis. These observable indicators were as follows, Selectivity, Stability and Precision.
Materials and Methods include Chromogenic Media CHM-Gn “A” and “B” obtained from two different manufacturers, Clinical Stool Specimens(Sterile Collected), VRE strains from culture collections (Table A) and Others: Microscopes, Saline, Agar Plates, Tubes, Incubator Inoculation of all Enterococcus species into two sets of media for
Selectivity determination. For control monitoring, positive control strains from VRE reference (n=4) from two different sources were also cultured. A dilution of two Enterococcus spp, Enterococcus faecium n=7; and Enterococcus faecalis ;n=2) reference and streaking on unadulterated and assorted cultures of both Media, followed by a 48-hour incubation. Comparative testing”(3). An enrichment step was incorporated for stool culture.
Results
Media “A” and “B” produced exemplary results after they were enriched overnight and incubated for 48-hours. Both media had a 98% (53/54) sensitivity, 95.5% (196/205) specificity. Both media revealed a typical colony appearance as described by the manufacturers (Table A). Comparative analysis revealed unacceptable results after foul bacteria were cultured, hence one of the VRE spp undetected from the two media. This was as a result of plating saline diluted stool samples on media “A” and “B” Comparable screening of over 200 stool specimens on media “A” and “B” was analyzed, resulting in a colonization index of 0.21(21%) for both media (Table A). The MLVA (Multiple Locus Variable Analysis) was used to determine VREfaecium’s genetic relations among its isolates(Over 40 isolates), where a summation of six unlike MLVA variety was arrived at (5). These varieties were expressed in both media.
Discussion
Both CHM-Gn media inhibited the cultivation of Enterococcus spp except one of its species. Generally, VRE cultivated better on “A” than on “B”. On medium “B”, the edges of minute colonies appeared dispersed with no noticeable margins due to the “cloudy” medium backdrop. (Table A) Hence, VRE colonies on “B” medium were further challenging, hence unnoticed for second round culturing process unlike VRE colonies on “A”. Pediococcus spp are frequently inoculated from stool samples(human), where patients having a history of chronic illnesses and plus patients having abdominal surgery history with those of abdominal history are mostly affected. (6, 7).
The experimentation clearly revealed that both CHM-Gn media don’t absolutely fulfill prospects considering VRE exposure, particularly when inoculating stool samples straight or following pre-dilution. Irresistible cultivation of diverse bacteria organisms and yeasts on media “A” and “B” hinder agar assessment while culturing stools samples directly. Stool samples are therefore unsuitable for straight inoculation onto both media.
Analysis of colony color “B” revealed colony color stability during the 24 and 48 hrs growth periods. This is however unexpected according to CHM-Gn “A” manufacturers. On “A” medium, the appearance of different color shades of VRE species further complicates the analysis. Regrettably, instituting the phase of enrichment slowed VRE identification. Fortunately, using either media presents a VRE cost-cutting screening technique for stool analysis, hence safety measures decreasing detrimental infection prevalence.
References
Deshpande, L. M., T. R. Fritsche, G. J. Moet, D. J. Biedenbach, and R. N. Jones. 2007.Antimicrobial resistance and molecular epidemiology of vancomycin-resistant enterococci from North America and Europe: a report from the SENTRY antimicrobial surveillance program.Diagn. Microbiol. Infect. Dis. 58:163–170.
Top, J., R. Willems, and M. Bonten. 2008. The emergence of CC17 Enterococcus faecium: from commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 52:297–308.
Delmas, J., F. Robin, C. Schweitzer, O. Lesens, and R. Bonnet. 2007. Evaluation of a new chromogenic medium, ChromID VRE, for detection of vancomycin-resistant Enterococci in stool samples and rectal swabs. J. Clin. Microbiol. 45:2731–2733.
Ledeboer, N. A., K. Das, M. Eveland, C. Roger-Dalbert, S. Mailler, S. Chatellier, and W. M.Dunne. 2007. Evaluation of a novel chromogenic agar medium for isolation and differentiation of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis isolates. J. Clin. Microbiol.45:1556–1560.
Top, J., L. M. Schouls, M. Bonten , and R. Willems. 2004. Multiple-locus variable-number tandem repeat analysis, a novel typing scheme to study the genetic relatedness and epidemiology of Enterococcus faecium isolates. J. Clin. Microbiol. 42:4503-4511.
Barton, L. L., E. D. Rider, and R. W. Coen. 2001. Bacteremic infection with Pediococcus: vancomycin-resistant opportunist. Pediatrics 107:775–776.
Mastro, T. D., J. S. Spika, P. Lozano, J. Appel, and R. R. Facklam. 1990. Vancomycin-resistant Pediococcus acidilactici: nine cases of bacteremia. J. Infect. Dis. 161:956–960.
Tables and Figures
Table a. Cultivated VRE and reference spp in media “A” and “B” Colonial appearance
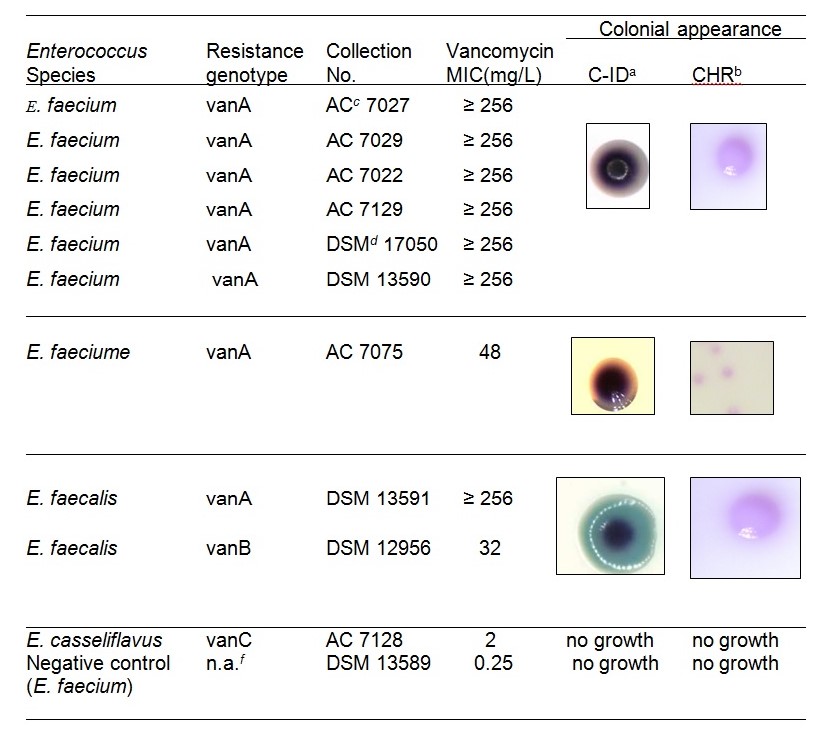
Table b. VRE stool analysis (n = >200) revealed cultivated in Both CHM-Gn media from similar vancomycin broth enriched media incubated for 48 hours
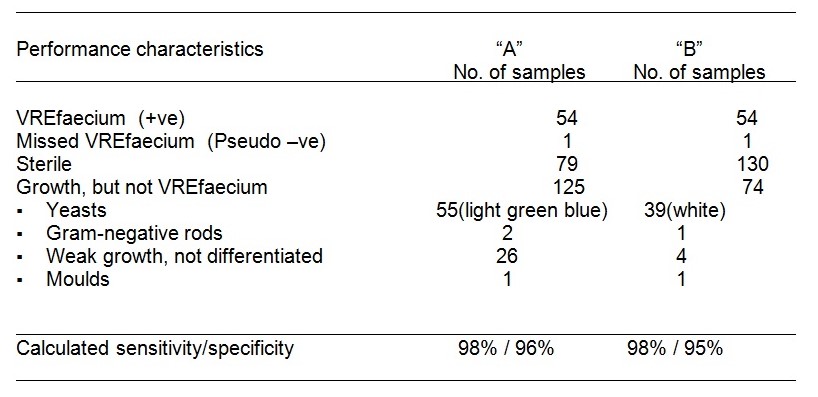
Table c. Multiple Locus Variable Analysis (MLVA) revealing MLVA variety and colony results of >40 VREfaecium, >200 isolates