Introduction
The current energy crisis has shifted focus from conventional fuels to renewable energy, including fuel cells. Fuel cells are electrochemical cells that transform the built-in chemical energy in fuels into electrical energy (Rahimnejad et al. 2015). Benefits of fuel cells include the absence of emissions that pollute the environment, high efficiency, and a lack of mobile parts, which eliminates noise pollution. However, their main shortcoming is the production of vast amounts of biomass. Microbial fuel cells (MFCs) are fuel cells that use active microorganisms as biological catalysts in anaerobic anode partitions to generate bioelectricity (Santoro et al. 2017). MFCs exploit the catabolic activity of bacteria to convert chemical energy into electricity. During the oxidation of chemicals, bacteria convey electrons to a progression of respiratory enzymes that store energy as adenosine triphosphate (ATP) inside the cell (Parkash 2016). The electrons are then discharged to an electron acceptor, which generates electrical energy. The purpose of this experiment was to determine the electrogenic potential of three different Enterobacteriaceae.
Materials and Methods
A single-chamber microbial fuel cell using nutrient agar as the proton exchange medium (PEM) was assembled in a biosafety cabinet, as described in the lab manual. 2 ml of Enterobacter aerogenes culture was added to the agar in the anodic compartment and allowed to set. The assembly of the MFC was then completed. The setup was incubated at 37oC. A similar procedure was repeated twice using Citrobacter freundii and Morganella morganii. The voltmeter readings in mV were taken at the end of the class and then seven days later. The data on electricity generated by the Enterobacteriaceae are recorded in Table 1.
The growth of E. aerogenes was determined indirectly in liquid medium in batch culture by measuring the absorbance at 550 nanometers at 15-minute intervals. The growth from two conditions (shaking and non-shaking) was determined and recorded in Table 2 and Figure 2.
Results
Rate of Voltage Generation
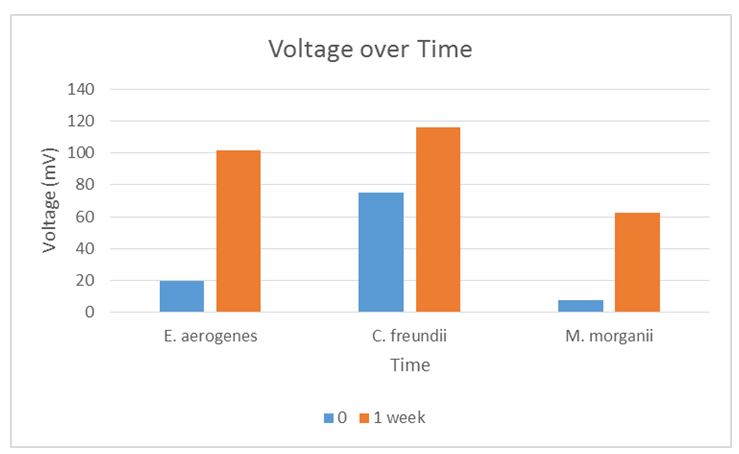
C. freundii had the highest voltage generation potential followed by E. aerogenes and M. morganii, as indicated in Table 1 and Figure 1.
Table 1: Voltage generation over time
Generation Time Data
The generation time of E. aerogenes was shortened by shaking.
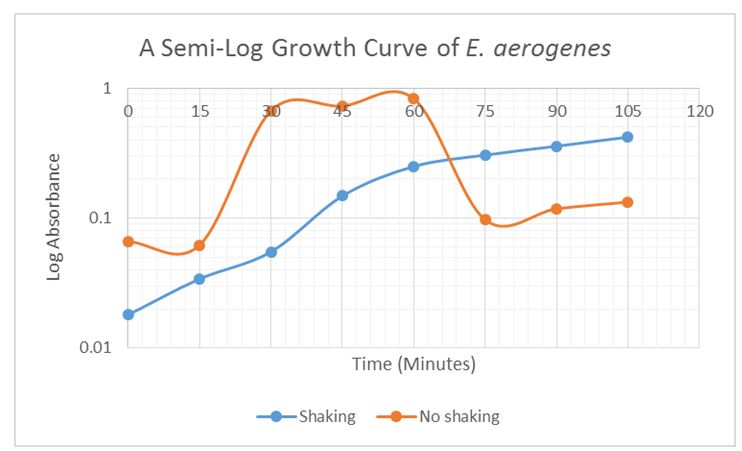
Table 2: Generation time data for E. aerogenes
Discussion
The findings were similar to those recorded in the literature. C. freundii is known to be the Enterobacteriaceae species with the highest electrogenic potential (Chen, Chen, & Chung 2014). E. aerogenes had the highest startup voltage, as also reported by Zhuang et al. (2011). E. aerogenes and M. morganii are facultative anaerobes whereas C. freundii is an aerobic bacterium. The generation time data of E. aerogenes was consistent with the research findings that shaking of bacterial cultures shortened the lag phase and led to higher optical densities (Munna et al. 2014). This information may be useful in bacterial identification by comparing the voltage generated by different bacteria at specified times. The electrogenic potential of bacteria is beneficial in other biotechnology applications such as bioremediation, wastewater treatment, and the production of prized commodities.
References
Chen, CY, Chen, TY & Chung, YC 2014, ‘A comparison of bioelectricity in microbial fuel cells with aerobic and anaerobic anodes,’ Environmental Technology, vol.35, no. 3, pp. 286-293.
Munna, MS, Tamanna, S, Afrin, MR, Sharif, GA, Mazumder, C, Kana, KS, Urmi, NJ, Uddin, MA, Rahman, T & Noor, R 2014, ‘Influence of aeration speed on bacterial colony-forming unit (CFU) formation capacity,’ American Journal of Microbiological Research, vol.2, no. 1, pp. 47-51.
Parkash, A 2016, ‘Microbial fuel cells: a source of bioenergy,’ Journal of Microbial & Biochemical Technology, vol. 8, pp. 247-255.
Rahimnejad, M, Adhami, A, Darvari, S, Zirepour, A. & Oh, SE 2015, ‘Microbial fuel cell as new technology for bioelectricity generation: a review,’ Alexandria Engineering Journal, vol. 54, no. 3, pp. 745-756.
Santoro, C, Arbizzani, C, Erable, B & Ieropoulos, I 2017, ‘Microbial fuel cells: from fundamentals to applications: a review,’ Journal of Power Sources, vol. 356, pp. 225-244.
Zhuang, L, Zhou, S, Yuan, Y, Liu, T, Wu, Z & Cheng, J 2011, ‘Development of Enterobacter aerogenes fuel cells: from in situ biohydrogen oxidization to direct electroactive biofilm,’ Bioresource Technology, vol. 102, no. 1, pp. 284-289.