Introduction
With the advent of advanced forms of microbiological analysis, the observation of microorganisms has progressed considerably. Modern equipment makes it possible to observe the lifestyle of bacteria with real-time light microscopes; however, in the natural environment, most microbes are colorless. The lack of coloration may be convenient for observing the internal components, but on a broad scale is more of a limitation. Pigmented staining procedures for microorganisms using organic dyes have been used to solve this problem (Petersen and McLaughlin, 2021). Meanwhile, staining fulfills a simplifying function for better observation of organisms and covers the need for their identification: physiological features of different cells lead to inherently different staining results. Based on their known reference patterns and the accurate picture of stained microorganisms, the task of their classification identification becomes solvable. In this report, the framework of three staining procedures for several bacterial and spore lines, covering the differential needs of microbiological analysis, is explored in detail.
Method
Gram Staining
The generally accepted technique for Gram staining consists of the sequential use of several dyes and the washing agent necessary to fix the color. A bacterial smear — B. megaterium and E. coli — was applied to a sterile labeled room temperature slide using a Bunsen burner loop. A few drops of crystal violet pigment (gentian violet) were added to the smear for 1-2 minutes, and after washing off the excess, 1-2 drops of Lugol’s iodine were added. 95% ethyl alcohol was applied to the bacterial smear in the next step for decolorization. Finally, safranin helped fix the new bacterial Gram pigment: all excess pigments were washed off with tap water. The bacterial preparation was prepared for microscopic observation.
Spore Staining
A drop of distilled water was placed on a sterile labeled slide using a Pasteur pipette. A B. megaterium colony was transferred to this drop using a sterile loop; the film was rapidly dried and fixed using a Bunsen burner. The bacterial culture was covered with a towel soaked in malachite green. The pigment was fixed by maintaining a steam bath over which a slide was placed for several minutes. The dyed towel was removed, and the excess unfastened pigment was washed off with running water. Safranin contrast staining was used for the cooled bacterial smear to ensure that the smear was fixed with new pigment. The bacterial preparation was prepared for microscopic observation.
Acid-Fast Staining
A bacterial mixture of E. coli and M. smegmatis was placed in a sterile, bijou-labeled test tube, after which the suspension was fixed by bringing the tube to a Bunsen burner. Red carbolic-fuchsin was then used to stain the bacterial suspension for several minutes, after which the excess pigment was washed off. A 1% sulfuric acid solution allowed some of the bacteria to discolor. Contrast staining with methylene blue was then used to discolor the fixation of the new pigment. The bacterial preparation was prepared for microscopic observation.
Results
Since the only option to identify bacteria by staining is to use a light microscope, the current experiment resulted in microphotographs of various preparations.
Discussion
Gram Staining
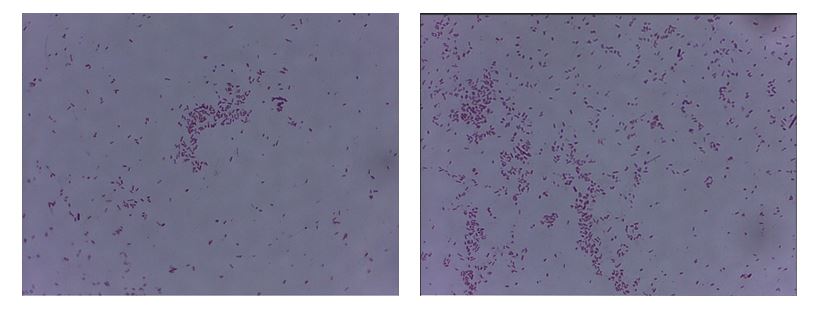
Bacterial staining solves many problems associated with simplified observation of cells using a microscope. When combined with microbial identification, it covers the challenges of microbiological laboratory analysis (Elga, 2021). Staining is differentially fixed on different microorganisms due to their physiological and morphological features. For example, Gram staining makes it possible to trace differences between Gram- and Gram+ bacteria with different cell wall structures. It is well known that the murein layer of the cell membrane of Gram+ bacteria is much thicker, which allows purple staining to initially fix, unlike Gram- bacteria, whose polysaccharide shell is much thinner. The current experiment showed that E. coli bacilli acquired a characteristic pink coloration, whereas the other cells were stained purple. The purple color corresponded to the Gram-positive bacteria B. megaterium, for which the pigment was initially fixed in the murein, in contrast to E. coli, which was unable to fix the gentian violet and was discolored with ethanol.
Spore Staining
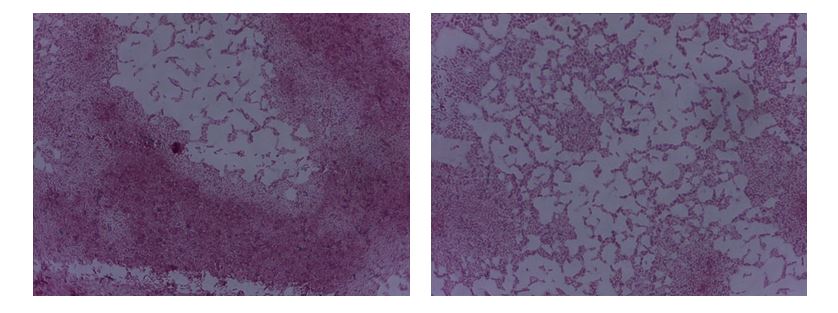
However, observation of bacteria can be complicated by unfavorable conditions affecting the organisms. In particular, bacteria can form endospores, which protect against external stimuli for the unfavorable season. Endospore staining is conducted using the spore staining technique developed by Dorner, Schaeffer, and Fulton (Aryal, 2018). During this staining, endospores turn bright green, whereas vegetative cells that do not form spores are stained with safranin. Reference to Figure 2 shows that most B. megaterium bacilli are intensely pink in color, whereas only trace amounts of globular spores are stained malachite green. Since B. megaterium can form endospores, it can be concluded that only a tiny fraction of the bacteria in the smear exist in this form (Lanzilli et al., 2016). This indicated that favorable conditions for the cells are present.
Acid-Fast Staining
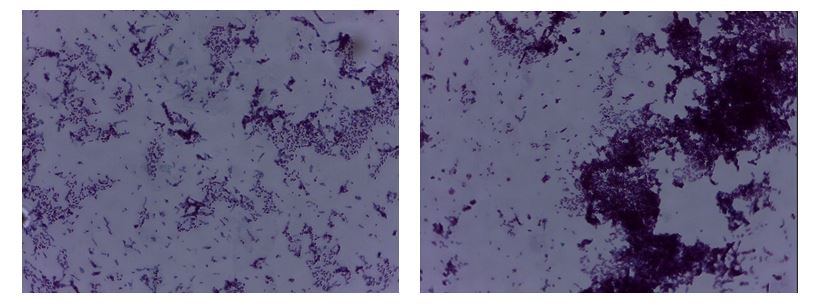
Finally, the acid-fast staining technique differentiates between acid-resistant and acid-unresistant bacteria. In other words, it is possible to use this method to identify those bacteria that can exist under extremely low pH conditions and do not respond to basic staining techniques, including Gram staining (Aryal, 2018). Using this method, acid-tolerant bacteria turn bright red or purple, whereas acid-tolerant strains fix the blue pigment. Reference to Figure 3 shows that most of the bacteria have purple coloration: these include M. smegmatis, which retained its original coloration (Burke, 2020). In contrast, E. coli recorded only blue pigment because it is an acid-resistant bacterium (SIL, 2019). In Figure 3, there are far fewer such blue prokaryotes, but it is also possible to distinguish them.
Reference List
Aryal, S. (2018) Acid-fast stain-principle, procedure, interpretation and examples. Web.
Aryal, S. (2018) Endospore staining-principle, reagents, procedure and result. Web.
Burke, K. C. (2020) Introduction. Web.
Elga (2021) Microbiology. Web.
Lanzilli, M., Donadio, G., Addevico, R., Saggese, A., Cangiano, G., Baccigalupi, L., Christie, G., Ricca, E. and Isticato, R. (2016) ‘The exosporium of Bacillus megaterium QM B1551 is permeable to the red fluorescence protein of the coral Discosoma sp.,’ Frontiers in Microbiology, 7(1752), pp. 1-8.
Petersen, J., and McLaughlin, S. (2021) Introduction to staining. Web.
SIL (2019) Acid-fast stain. Web.