Summary
Respiratory tract infections represent the group of infections that cause diseases of the throat, lungs, or airways, depending on the type. Thus, respiratory tract infections are divided into upper respiratory tract infections (URTIs), which cause diseases of the throat, nose, pharynx, and larynx, and lower respiratory tract infections (LRTIs), which cause diseases of the bronchi, trachea, and lungs (Kateete et al. 2010). LRTIs are usually associated with such serious conditions as pneumonia and bronchitis. LRTIs are usually viral in origin, and the pathogens that cause pneumonia and bronchitis include S. pneumonia, H. influenza, M. catarrhalis, S. aureus, and Klebsiella pneumonia (Pesavento et al. 2010). As a result of infections, patients have problems with breathing, and they report having fever and cough among other symptoms. The acute forms of these conditions can cause even the patient’s death if the infection is not treated appropriately (Shrestha, Pokhrel & Mohapatra 2009). In its turn, influenza can affect both URTIs and LRTIs. The H5N1 subtype of the influenza virus can have the most serious negative impact on the patient’s lungs and bronchi (Mishra & Bayer 2013).
In this case, the 59-year-old male has the signs of bronchitis. It is important to note that bronchitis is categorized as the LRTI, and the treatment depends on the pathogen that causes the condition. From this point, further microbiological investigation is necessary because it is significant to determine which bacterium causes bronchitis in the patient and prescribe the appropriate antibiotics to address it while referring to the antibiotics sensitivity testing. In spite of the fact that bronchitis can be caused by a variety of bacteria, the microbiological investigation allows determining the specific pathogen causing the disease development in the concrete patient, and the expected outcome is the discussion of the bacterium and identification of antibiotics to treat the disease effectively.
Materials and Methods
In order to conduct the microbiological investigation, the following materials were used:
- The sputum sample.
- Three agar plates: Horse blood agar (HBA), MacConkey agar (MAC), and Chocolate agar (CHA).
- Gram staining test.
- Catalase test.
- Coagulase test.
- Deoxyribonuclease (DNase) plate with hydrochloric acid.
- Antimicrobial susceptibility test discs.
The first step in the microbiological investigation was the culturing of the patient’s sputum sample characterized as a heavy purulent one with the help of three agar plates: HBA, CHA, and MAC. The sputum was cultured and incubated for 24 hours under aerobic condition (CO2). These plates were selected because according to Cain (2014), the use of HBA, CHA, and MAC plates contributes to the rapid growth of colonies of bacteria that can be further examined individually. Adegoke and Okoh (2014) also note that under aerobic conditions, many bacteria not only survive but also grow actively.
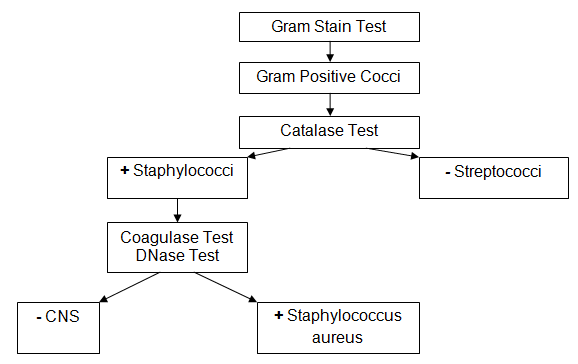
In 24 hours, the colonies from the three plates were examined to determine the morphology type. The colonies from the HBA plate were taken with the help of a sterile loop in order to conduct the Gram staining test. The Gram stain is important in order to identify the morphology of the bacterium or its type (Kaasch et al. 2014). The same colonies were used in order to conduct the catalase test for the purpose of distinguishing between Staphylococcus and Streptococcus. The catalase test is selected because Staphylococci usually have the enzyme catalase when it is not presented in Streptococci (Cain 2014; Chatterjee et al. 2009).
The next step was to conduct the coagulase test for the purpose of distinguishing between types of Staphylococci. The slide test was conducted in order to identify how the determined Staphylococcus can clot the plasma in order to identify Staphylococcus aureus. To confirm the presence of Staphylococcus aureus, it was reasonable to conduct the DNase test. This test is used for the detection of Staphylococcus aureus to prove the results of the previous tests because Staphylococcus aureus contains such enzyme as DNase (Wiriyachaiporn et al. 2013). The DNase plate was incubated for 24 hours.
When all tests were performed, it was necessary to conduct the antibiotics sensitivity test with the help of antimicrobial susceptibility test discs in order to determine to which types of antibiotics Staphylococcus aureus can be susceptible. Penicillin, gentamicin, chloramphenicol, oxacillin, cefoxitin, and erythromycin were selected for this test because of the high-level resistance of Staphylococcus aureus to many types of antibiotics (Sudhanthiramani, Swetha & Bharathy 2015). The results of the conducted tests are presented in the following section.
Results
The results of testing using HBA, CHA, and MAC plates under aerobic conditions indicate the presence of white and yellow colonies for HBA, white and yellow colonies for CHA, and pinky colonies for MAC (Table 1).
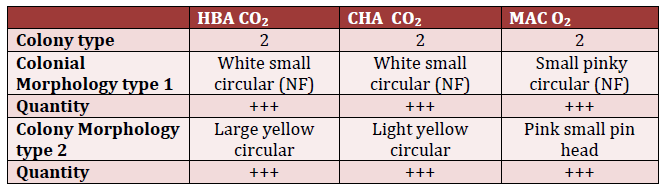
HBA – Horse blood agar.
MAC – MacConkey agar.
CHA – Chocolate agar.
NF – nuclear factor.
For the colonies from the HBA plate, the Gram staining test was conducted. It was found that Gram-positive cocci were presented in chains of three or four colonies or clusters similar to chains. The catalase test performed after the Gram test identified the enzyme catalase-positive bacterium. As a result, it was possible to speak about the presence of Staphylococci in the patient’s sputum sample. The coagulase test also presented a positive results, and the identified Staphylococcus was categorized as Staphylococcus aureus. This conclusion was also supported by the positive results of the DNase test because the clear zone and DNase +ve colonies were found (Table 2).
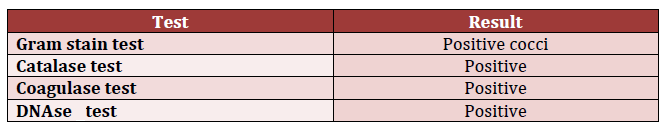
DNase – deoxyribonuclease.
The results of the Gram stain test allowed the development of the scheme for further microbiological investigation in order to confirm that the identified pathogen was of the Staphylococcus type (Figure 1).
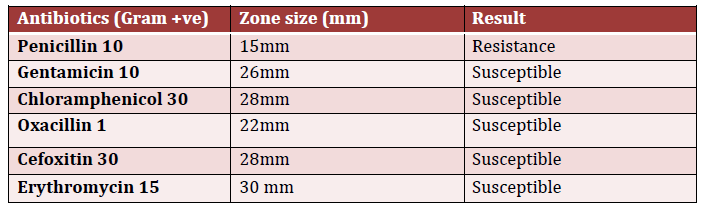
The antibiotics sensitivity test indicated that the identified Staphylococcus aureus was resistant to penicillin, but it was susceptible to gentamicin, chloramphenicol, oxacillin, cefoxitin, and erythromycin (Table 3).
Laboratory Report
Sample Description: Sputum sample.
Notes: Purulent sputum sample, possible bronchitis.
Date: 23/4/2016.
Gram: Gram-positive cocci, chains of colonies.
Culture: Growth of Staphylococcus aureus.
Comments: Susceptible to gentamicin, chloramphenicol, oxacillin, cefoxitin, and erythromycin.
Discussion
Bronchitis can be provoked by S. aureus, S. pneumonia, H. influenza, and M. catarrhalis among other bacteria. In 59-year-old patients, as well as elderly patients, the risks of developing complications associated with non-treated bronchitis increase, and it is important to identify the bacterium that causes the disease in the particular case (Kitara et al. 2011). Acute bronchitis is frequently caused by Staphylococcus aureus when the bacteria pass from the pharynx to bronchi, and the organism’s reaction to the bacteria is the inflammation with the production of mucus (Wiriyachaiporn et al. 2013). In order to identify whether bronchitis is caused by Staphylococcus aureus or other bacteria, it is necessary to examine the sputum sample and conduct a variety of tests.
In this case, the Gram stain test indicated that the bacteria were Gram-positive cocci that are typical of both Staphylococcus and Streptococcus, and they resembled both clusters and chains of colonies. Therefore, additional testing was necessary to distinguish between Staphylococcus and Streptococcus. The catalase test indicated that the bacteria are catalase–positive, and this condition is typical of Staphylococcus rather than Streptococcus. Moreover, this condition is characteristic of Staphylococcus aureus. In order to support the assumption about the morphology of the bacterium, it was necessary to conduct the coagulase and DNase tests. These tests allowed speaking about the bacteria as coagulase-positive. They contributed to creating clear zones. Therefore, the conclusion was that the male patient’s bronchitis was caused by Staphylococcus aureus.
In the 59-year-old patient, Staphylococcus aureus can also cause pneumonia; therefore, it was necessary to conduct the antibiotics sensitivity test in order to identify antibiotics that are most appropriate to be used in the patient’s case. Mishra and Bayer (2013) state that individual sensitivity can influence the effectiveness of using gentamicin, chloramphenicol, oxacillin, cefoxitin, and erythromycin in order to treat bronchitis in elderly patients. From this point, it is important to note that patients aged 59 years old and older are in the group of people who are usually affected by Staphylococcus aureus because of the lowered immunity (Kitara et al. 2011).
Public health considerations related to the treatment of persons with Staphylococcus aureus include the prescription of appropriate antibiotics. It is also important to focus on the isolation of patients during the period of treatment and the decrease of risks associated with the further possible infecting or spread of the untreated Staphylococcus aureus in the organism. Much attention should be paid to avoiding the auto-infection and monitoring the symptoms in the patient during the treatment.
Reference List
Adegoke, A & Okoh, 2014, ‘Species diversity and antibiotic resistance properties of Staphylococcus of farm animal origin in Nkonkobe Municipality, South Africa’, Folia microbiologica, vol. 59, no. 2, pp. 133-140.
Cain, H 2014, Microbiological laboratory techniques manual, University of Melbourne, Melbourne.
Chatterjee, S, Ray, P, Aggarwal, A, Das, A & Sharma, M 2009, ‘A community-based study on nasal carriage of Staphylococcus aureus’, Indian Journal of Medical Research, vol. 130, no. 6, pp. 742-748.
Kaasch, A, Barlow, G, Edgeworth, J, Fowler, V & Hellmich, M 2014, Staphylococcus aureus bloodstream infection: a pooled analysis of five prospective, observational studies’, Journal of Infection, vol. 68, no. 3, pp. 242-251.
Kateete, D, Kimani, C, Katabazi, F & Okeng, 2010, ‘Identification of Staphylococcus aureus: DNase and Mannitol salt agar improve the efficiency of the tube coagulase test’, Annals of Clinical Microbiology and Antimicrobials, vol. 9, no. 1, 1-10.
Kitara, L, Anywar, A, Acullu, D, Odongo-Aginya, E & Aloyo, J 2011, ‘Antibiotic susceptibility of Staphylococcus aureus in suppurative lesions in Lacor Hospital, Uganda’, African Health Sciences, vol. 11, no. 3, pp. 34-39.
Mishra, N & Bayer, 2013, ‘Correlation of cell membrane lipid profiles with daptomycin resistance in methicillin-resistant Staphylococcus aureus’, Antimicrobial Agents and Chemotherapy, vol. 57, no. 2, pp. 1082-1085.
Pesavento, G, Ducci, B, Comodo, N & Nostro, 2010, ‘Antimicrobial resistance profile of Staphylococcus aureus isolated from raw meat: a research for methicillin-resistant Staphylococcus aureus (MRSA)’, Food Control, vol. 18, no. 3, pp. 196-200.
Shrestha, B, Pokhrel, B & Mohapatra, T 2009, ‘Phenotypic characterization of nosocomial isolates of Staphylococcus aureus with reference to MRSA’, The Journal of Infection in Developing Countries, vol. 3, no. 7, pp. 554-560.
Sudhanthiramani, S, Swetha, C & Bharathy, S 2015, ‘Prevalence of antibiotic-resistant Staphylococcus aureus from raw milk samples collected from the local vendors in the region of Tirupathi, India’, Veterinary World, vol. 8, no. 4, pp. 478-481.
Wiriyachaiporn, S, Howarth, P, Bruce, K & Dailey, L 2013, ‘Evaluation of a rapid lateral flow immunoassay for Staphylococcus aureus detection in respiratory samples’, Diagnostic Microbiology and Infectious Disease, vol. 75, no. 1, pp. 28-36.