Introduction
“Affinity chromatography is a method used to separate biochemical mixtures and is based on highly specific biological interaction between an enzyme and substrate, antigen and antibody” (Melander et al. 1999) This technique is utilized in; Purification and concentration of substances from a blend into a buffering solution, to cut down the amount of one constituent of a solution and in the investigation the different biological compounds that bind to specific substances. (Melander et al. 1999) This paper seeks to review the affinity column method used in laboratory investigations.
Specifically, the method and its principle will be outlined and three studies analyzed to underscore the importance of the method, various challenges, and how they are being improved.
Technique description
Principle
Chromatography often goes through the immobile and mobile phases. The immobile phase is provided by a gel matrix, usually an Agarose gel; a linear sugar molecule sourced from algae (Mattiasson et al. 1999). During the process, heterogeneous molecules having specific properties will be separated from other molecules that lack the targeted property. In essence, the target molecules are trapped in the solid medium (Mattiasson et al. 1999). The process then goes into the second phase (mobile) in which the mixture is washed and the target molecule eluted (Melander et al. 1999).
The binding of the molecules to a solid phase can be done using the column chromatography in which the “solid medium is packed into the column and the initial mixture run through the column to allow setting.” (Melander et al. 1999) Awash buffer is run through the column, the elution buffer applied, and then collected from the column. Alternatively, this can also be achieved by a newer method that utilizes the advantages of batch setup; this method is referred to as expanded bed adsorption.
In this technique, “the solid phase particles are placed in a column then the liquid phase is pumped from the bottom to exit from the top” (Mattiasson et al. 1999). Gravitational force is exploited in this setup and it ensures that the solid phase remains with the particles as the liquid phase exits from the column (Melander et al. 1999).
Affinity chromatography is utilized in various operations which may include; “nucleic acid purification, protein purification from cell-free extracts and purification from blood” (Melander et al. 1999). The technique is more often used in medical laboratory investigations to purify antibodies from blood serum. In this case, the Agarose gel may contain specific antigens which covalently attach to the antibodies (Melander et al. 1999).
The same can be achieved using the” immobilized metal ion affinity chromatography (IMAC)” (Mattiasson et al. 1999). This method is based on the particular covalent bonds that exist between amino acids, specifically histidine and metals (Mattiasson et al. 1999). Separation is usually achieved when the “proteins that have an affinity for metal ions are retained in a column containing the metal ions” (Melander et al. 1999). The metal ions that are commonly used include gallium, cobalt, zinc, nickel, iron, copper among others.
Purification of recombinant proteins can be regarded as the most frequent application of affinity chromatography (Melander et al. 1999). The proteins in question may have been passed through genetic modification to allow optimum selection for affinity binding. In this case, the proteins are referred to as “fusion proteins” (Mattiasson et al. 1999). Various types of tags are available for use; the most common types include “glutathione-S-transferase (GST), hexahistidine (his), and maltose-binding protein (MBP)” (Melander et al. 1999). The tags are often used in the elution process. For instance, “the GST has an affinity for glutathione which is commercially available as immobilized glutathione agarose” (Mattiasson et al. 1999). During the elution process, excess glutathione is used to displace the tagged protein.
Studies conducted to improve affinity chromatography technique
The application of the affinity column technique has had many challenges. The issues revolve around the specificity and sensitivity of the eluted fractions, which in most cases are proteins (Arakawa et al. 2007). Proteins are prone to degradation when subjected to physical and chemical processes. The challenge often lies in developing affinity column techniques that will allow the maximum collection of the target proteins with no or minimal effects (Mattiasson et al. 1999) Various studies have been conducted to establish how affinity chromatography can be improved for effective use in protein elution and quantification (Melander et al. 1999). The following is a review of various studies conducted to improve protein recovery.
Improved performance of column chromatography by arginine: Dye-affinity chromatography (Arakawa et al. 2007)
A study was carried out by Arakawa et al to verify the improvement in the performance of column chromatography when arginine is utilized in the dye affinity chromatography (410). The dye-affinity column chromatography provides a faster and more convenient protein purification protocol. This is because proteins usually have a high affinity for dye columns (Arakawa et al. 2007). In addition, a bigger proportion of samples can be eluted without preceding treatment procedures. In this method, “covalently attached group-specific ligands are used for purification of enzymes, such as dehydrogenases and kinases” (Arakawa et al. 2007).
Columns with blue dye adhere to enzymes that need “adenyl-containing cofactors such as NAD and NADP. Lactate dehydrogenase (LDH) binds to blue dye columns and the bound enzyme is eluded by salts, but with low recovery and broad elution peak ” (Arakawa et al. 2007). In this study, arginine was investigated to find out whether it can solve these challenges. “Thus its ability to increase collection and resolution using Blue-Sepharose and LDH as a model protein” (Melander. et al. 1999)
The results of the study revealed that the usual eluent, “NaCl indicated a wide elution peak with a low recovery of lactate dehydrogenase, at most approximately 60 using 2M salt The recovery was seen to decrease with either increase or decrease in the NaCl concentration” (Arakawa et al. 2007) However, when arginine was used in place of NaCl, the recovery of lactate dehydrogenase was higher than 80% at more than 0.5 M (Arakawa et al. 2007). The recovery was almost free of the arginine concentration used (Mattiasson et al. 1999). This resulted in a “sharper elution peak, leading to the recovery of a protein solution of good concentration” (Arakawa et al. 2007).
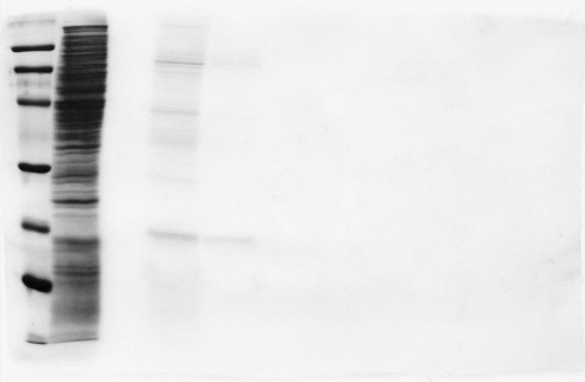
Lane 1 (molecular weight marker), lane 2 (Xow through), lane 3 (buVer wash), lane 4 (0.5 M arginine), lane 5 (second 0.5M arginine), lanes 6 and 7 (1Marginine) and lanes 8 and 9 (2 M arginine). The band at 21 kDa corresponds to NDK, as examined by Western blotting analysis using an anti-NDK(Ha293) antibody (MI and MT, unpublished results). (Arakawa et al. 2007).
A new Application of imprinted Polymers: Specialization of organotin compounds (Gallego-Gallegos et al. 2010)
This study was carried out by Gallego-Gallegos et al. to determine the application of molecular imprinting technology in the preparation of a chromatographic stationary phase of specific affinity for speciation purposes. The study aimed to develop a simple, selective, rapid, and sensitive method for the quantitative determination of organotin compounds in the environment. Speciation for this type of compound is usually done using gas chromatography, because of good resolution and sensitivity (Mattiasson et al. 1999). However, organotin compounds (OTCs) have poor volatility and thus often require the initial derivatization step (Gallego-Gallegos et al. 2010).
This is not usually the case in liquid chromatography. In this study, Tributyltin was chosen as the template and a noncovalent technique was applied (Melander et al. 1999) Evaluation of three polymerization methods was done, the three methods include; “a composite material, a polymer prepared via-inverter grafting and an emulsion polymer” (Gallego-Gallegos et al. 2010). The results of the study showed that the grafted inverter and the emulsion polymers formed cavities (Mattiasson et al. 1999). The “emulsion polymers achieved a resolution of four organotin compounds” (Melander et al. 1999). The study revealed that this “stationary phase led to good recovery for all the species that were tested. The high selectivity prevented matrix interferences” (Gallego-Gallegos et al. 2010).
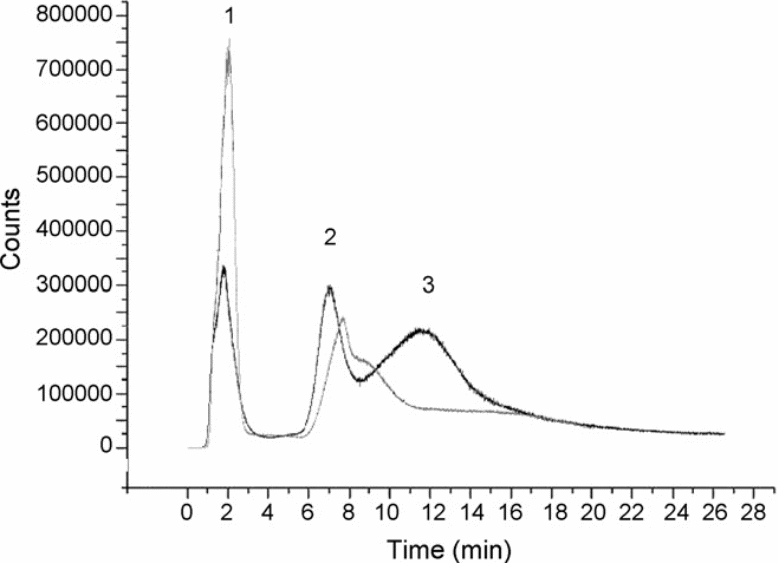
Monitored isotope 120Sn; loading 20 ng of each compound in water; injection loop: 0.2 mL; mobile phase flow: 0.5mLmin−1. Mobile phase gradient applied: (1) 0–2 min 100% MeOH; (2) 2–6 min: move to HAc 10% + 0.5% TEA in MeOH; (3) 6–30 min HAc 10% + 0.5% TEA in MeOH. Peaks: 1. Non-specific bonding of all OTCs; 2. TBT; 3. TPhT + DBT (Gallego-Gallegos et al. 2010).
Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers (Whiteaker et al. 2007).
This study was carried out by Whiteaker et al. to investigate the use of antibody based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers (44). The study was prompted by the challenge that is often faced in the development of highly sensitive and specific assays for quantifying proteins (Whiteaker et al. 2007). In this study, the researchers “optimized a magnetic bead-based platform to high-throughout peptide capture and demonstrated that antibody capture followed by mass spectrometry can achieve ion signal enhancement on order of 1000, sufficient for quantifying biomarkers in the physiologically relevant mg/ml range” (Whiteaker et al. 2007).
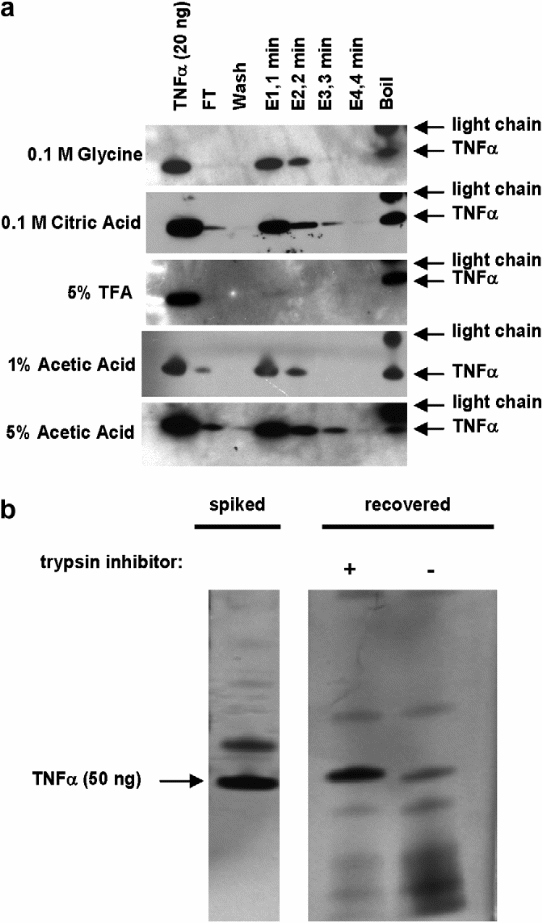
- Western blot (cropped) showing recovery of 50 ng (2940 fmol) TNFa spiked into 1 mL human serum, captured at 4 _C overnight by 20 lL of anti-TNFa-polyclonal-antibody-coated beads. ‘‘FT’’ refers to flow-through. ‘‘Wash’’ refers to four times washing of beads with 1 mL of 0.5M NaCl. Multiple elution buffers were tested, as indicated. Four aliquots of elution buffer (labeled E1–E4, 8 lL each) were added to 20 lL of antigen-bound antibody-coated beads at RT in 1-min intervals (total of 4 min). At the end of the elution period, the beads were boiled in SDS buffer to remove residual protein (‘‘Boil’’). Note that the secondary antibody used is a goat antimouse antibody, which reveals mouse lightchain fragments released by boiling.
- SDS–PAGE of antibody enrichment of 50 ng of recombinant TNFa spiked into 400 lL of human serum. The gel image shows recovery of TNFa from a trypsin digest of human serum with (+) and without (-) addition of trypsin inhibitor following the digestion (prior to the antibody-capture step ( Whiteaker et al. 2007).
Conclusions
From the studies above it has been shown that the affinity column performance can be enhanced by improving different components such as the stationary phase, the eluent, and binding capabilities (Melander et al. 1999) However, the improvement is only tailor made for specific applications. More studies need to be carried out to determine how the overall affinity chromatography technique can be enhanced to achieve optimum recovery of separated fractions (Melander et al. 1999).
References
Arakawa T, Ejima D, Tsumoto K, Ishibashi M, Tokunaga M (2007) Improved performance of column chromatography by arginine: dye-affinity chromatography. Protein Expr Purif 52: 410–414.
Mercedes Gallego-Gallegos, Maria Liva Garrido, Riansares Munoz Olivas, Patricia Baravalle, Claudio Baggiani, Carmen Camara (2010). A new application of imprinted polymers: Speciation of organotin compounds. Journal of Chromatography 1217: 3400–3407.
Whiteaker, J. R., Zhao, L., Zhang, H. Y., Feng, L., Piening, B. D., Anderson, L., and Paulovich, A. G. (2007) Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Anal. Biochem. 362, 44–54.
Mattiasson B, Galaev IYu & Garg N (1999) Polymer-shielding dyeaffinity chromatography. J. Mol. Recogn. 9: 509–514.
Melander, Horvath et al. (1999) Column Affinity Chromatography Biochem 200-215.