Introduction
Oshima et al (1987) data claim Beta-glucuronidase (figure 1) is a biological enzyme. Oshima et al (1987) findings and Bernfeld and Fishman’s (1953) data suggest that beta-glucuronidase hydrolyses conjugated glucuronides. Human Beta-glucuronidase hydrolyses dermatan and keratan sulfates. This catalytic reaction is represented by the word equation below
Beta-D-glucuronoside + H2O = D-glucuronate + an alcohol
Gloser et al (1975) findings documents that Beta-glucuronidase is encoded by the GUSB gene. Beta-glucuronidase has wide application in the determination of urinary steroids as documented by Bernfeld and Fishman (1950; 1953) data. Beta-glucuronidase hydrolyses glucuronide pro-drugs into active drugs as proposed by Bernfeld and Fishman (1950) and verified by Bernfeld and Fishman (1953) data and Oshima et al (1987) findings. The use of Beta-glucuronidase in hydrolysis is characterized by a very slow reaction which has contributed to its limited therapeutic application.
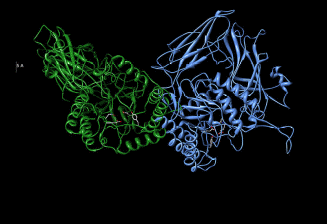
Characteristics of human β-glucuronidase
Contractor and Shane’s (1972) data claim that human β-glucuronidase has a molecular weight of 72532.58 daltons. Similar findings have been documented by Glaser et al (1975) findings. β-glucuronidase is composed of a sialic acid (Oshima et al, 1987) that is attached to a glycoprotein. Buehler et al (1951) data suggest that β-glucuronidase exhibits chemical heterogeneity that is brought about by different electron densities of different electron-withdrawing groups and electron-donating groups that form β-glucuronidase active sites as illustrated by figure 1. Buehler et al (1951) data have been duplicated by Christner et al (1970) studies.
In literature, β-glucuronidase has been documented to have an optimum PH of 4.4 (Dean, 1974; Oshima et al, 1987). Other studies have documented that β-glucuronidase has an extinction coefficient of 17 and an isoelectric point that measures 5.1 (Christner et al, 1970; Bernfeld and Fishman, 1953). β-glucuronidase has been determined to have the capacity to hydrolyze conjugated glucuronides. β-glucuronidase cannot hydrolyze alpha-glucosides or beta-glucosides due to its stereochemistry (Oshima et al, 1987).
Organic peroxides have been documented to have the capacity to inhibit β-glucuronidase hydrolysis by binding on its active sites as illustrated by figure 1 which locks active sites from accessing electron sinks (Christner et al, 1970). β-glucuronidase has been documented to have a stability of 6-12 months under thermo conditions characterized by a 2-8 degree Celsius range.
The assay of β-glucuronidase
The purification protocol of β-glucuronidase adopts a method that was described by Bernfeld and Fishman (1950; 1953). The purification protocol uses phenolphthalein glucuronidated as a substrate.
Quantities for the method
The purification protocol recommends the use of one Worthington unit equivalent of the reagents. Therefore one Worthington unit of β-glucuronidase would require Worthington equivalent of phenolphthalein glucuronidated for complete hydrolysis. One Worthington is equal to 19,000 Fishman units.
Conditions for the reaction
The purification protocol should be carried out at 37 degrees Celsius at a PH of 4.5.
Reagents required for the hydrolysis
The purification protocol requires the following four reagents

Requirements for the β-glucuronidase reaction
β-glucuronidase should be dissolved at a concentration of 2mg/ml in cold sodium chloride whose concentration should be 0.15 Molar. The β-glucuronidase should be standardized to 2mg/ml or a concentration range of 0.25mg/ml to 2.0mg/ml is acceptable.
The procedure for the process
- The water bath should be incubated at 37 degrees Celsius for five minutes before carrying out the purification protocol. This is meant to thermoregulate the temperature.
- Number the pipettes for the reaction
- One pipette should be left blank
- Into each of the numbered pipettes, 0.1ml of enzyme β-glucuronidase should be added. The blank pipette should be taken as control and water should be added.
- One of the pipettes should contain a 0.1 molar solution of Sodium Acetate whose volume should be 0.8ml
- The other pipette should contain 0.1 ml of o.01 molar solution of Phenolphthalein glucuronidated.
- The reaction should be incubated for 30 minutes at 37 degrees Celcius.
- The reaction should be stopped at 30 minutes and 5.0 ml of 0.2 molar solution of glycine added. This should be followed by a 0.2 molar solution of sodium chloride.
- The reagents should then be removed from the water bath and cooled.
- The value of A540 against the blank should be noted down.
- Separation should then be carried out through Thin-layer chromatography (TLC).
- This should be followed by X-ray diffraction, carbon-14 Nuclear magnetic resonance (C-NMR), or Hydrogen-nuclear magnetic resonance. (H-NMR)
The activity of β-glucuronidase fusion protein is determined by adding 0.5 mM of 4-methylumbelliferyl β-D-glucuronide in a beta-glucuronidase reaction buffer (50 mM Bis-Tris, 50 mM triethanolamine, 100 mM acetic acid, 100 ng/ml bovine serum albumin, pH 7.0) for 30 minutes at 37° C thermostated conditions. The reaction is terminated by adding an equal volume of stop buffer (1 M glycine, 0.5 M sodium bicarbonate, pH 11). The fluorescence of 4-MU is measured at excitation/emission wavelengths of 355/460 nm in a Gemini EM microplate spectrofluorometer (Molecular Device, Sunnyvale, Calif.).
β-glucuronidase which belongs to the family of organic compounds known as the glucuronidase family hydrolyses glycosaminoglycans to produce non-reducing end β-D-glucuronic acid residues. The Amino acid sequence of the precursor which represents human β-glucuronidase is illustrated below.

The mature form of Human beta-glucuronidase
Based on Human beta-glucuronidase [BGLR_HUMAN.organism: Homo sapiens (human)], the Human beta-glucuronidase has an amino acid sequence of 651. The secondary structure for the Human beta-glucuronidase is however complex as illustrated by figure 1.
The signal peptide has a sequence of 22 amino acids (1-22). The mature form of BGLR_HUMAN.organism: Homo sapiens (human)] has a sequence of 629 amino acids (23-651).
The active site of BGLR_HUMAN.organism: Homo sapiens (human)] occurs at 451st position of the BGLR_HUMAN.organizm: Homo sapiens (human)].
Modification of BGLR_HUMAN.organizm: Homo sapiens (human)
The modification of BGLR_HUMAN.organism: Homo sapiens (human)], can be carried out at the following positions of BGLR_HUMAN.organizm: Homo sapiens (human)], at 173, 272, 420, and 631. All modifications for the BGLR_HUMAN.organism: Homo sapiens (human)] can be achieved through glycosylation. The glycosylation at the proposed positions of BGLR_HUMAN.organism: Homo sapiens (human)], occur through N-linked terminal.
Conflicts in the BGLR_HUMAN.organizm: Homo sapiens (human)
There occur structural conflicts on BGLR_HUMAN. organism: Homo sapiens (human)] at the following positions namely 14 (from Q to E), 21 (from V to L), and 487 (from M to L).
Catalytic activity
The catalytic activity of beta-glucuronidase occurs through hydrolysis. Hydrolysis of beta-glucuronidase results in alcohol and beta-D-glucuronoside as represented by the word equation below.
Beta-D-glucuronoside + H2O = D-glucuronate + an alcohol.
Main inhibitor of beta-glucuronidase
The primary inhibitor of beta-glucuronidase is L-aspartic acid. This results from similarity.
The sub-unit structure of beta-glucuronidase
The subunit structure of beta-glucuronidase is homo-tetramer.
The ontology of human beta-glucuronidase
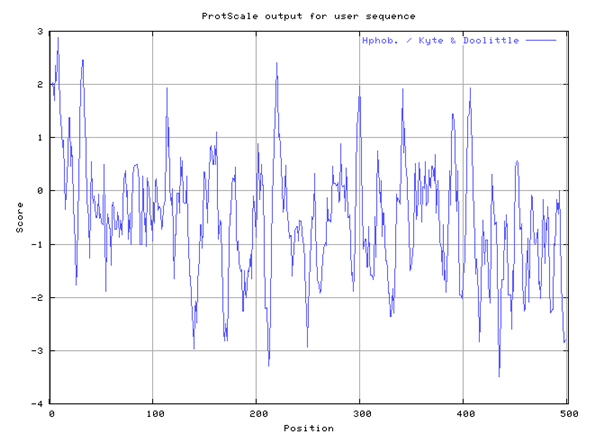
The ontology of beta-glucuronidase is characterized by its sugar-binding property, its action binding property as that defines the beta-glucuronidase molecular function, and its reactivity. The sugar-binding property of beta-glucuronidase results from beta-glucuronidase selective interaction non-covalently due to its hydrophilic properties (figure 2) with carbohydrates that are characterized by the presence of mono-, di or tri active sites.
The cation binding property of beta-glucuronidase is characterized by beta-glucuronidase selective interaction non-covalently with cations which implies, the activity of beta-glucuronidase is based on its nucleophilic property as opposed to its electrophilic property. The nucleophilic property of beta-glucuronidase is brought about by the presence of electron-donating groups that increase electron density hence making beta-glucuronidase active sites act as electron sources to a favored electron sink that electron-withdrawing groups that decrease electron density or electron sinks are attached to a better leaving group. Beta-glucuronidase activity is characterized by a catalysis reaction that is a subset of hydrolysis.
Beta-D-glucuronoside + H2O = an alcohol + D-glucuronate.
The Human beta-glucuronidase has many sites of hydrophobic and hydrophilic ends as illustrated by the Kyte-Doolittle hydropathy plot in figure 2.
Human beta-glucuronidase mature form sequences
The amino acid sequence for a mature form of beta-glucuronidase is represented in table 2.
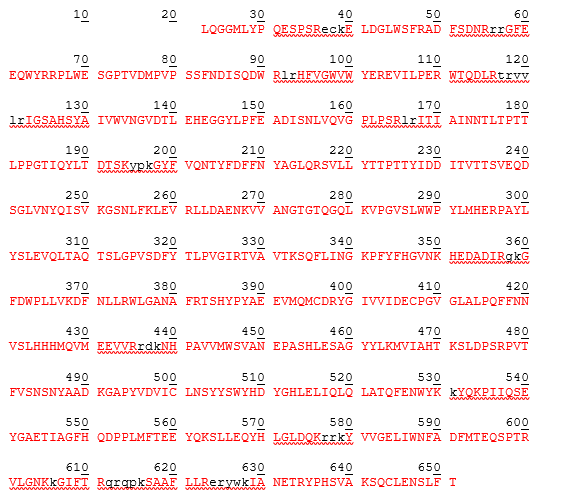
The human beta-glucuronidase mature form is represented by amino acid sequence from positions 23-651 as demonstrated by table 3 below. The theoretical pI is determined to be 6.44 while the molecular weight has been calculated to be 72578.35 while the monoisotopic molecular weight has been calculated to 72532.58.
Table 3: different masses of human Beta-glucuronidase at different amino acid sequence positions
Due to mutation, variation in Beta-Glucuronidase has been documented in the literature. These variations occur at position 30 (P to S), position 38 (C to G), position 52 (S to F), position 136 (G to R), position 148 (P to S), position 150 ( E to K), position 152 ( D to N), position 176 (L to F), position 216 (R to W), position 243 (L to P), position 320 (Y to C), position 320 (Y to S), position 339 (N to S), position 350 (K to N), position351 (H to Y), position 354 (A to V), and many other variations.
Individual amino acid values
Ala: 1.800
Arg: -4.500
Asn: -3.500
Asp: -3.500
Cys: 2.500
Gln: -3.500
Glu: -3.500
Gly: -0.400
His: -3.200
Ile: 4.500
Leu: 3.800
Lys: -3.900
Met: 1.900
Phe: 2.800
Pro: -1.600
Ser: -0.800
Thr: -0.700
Trp: -0.900
Tyr: -1.300
Val: 4.200: -3.500: -3.500: -0.490
Hydropathy plot maximum and minimum values
Minimum value = -3.503
Maximum value = +2.866
Based on the hydropathy plot, Beta-Glucuronidase exhibits hydrophobic and hydrophilic properties. Beta-Glucuronidase in practice tends to be more hydrophobic due to the influence of a high minimum value of -3.503 on the hydropathy plot compared to +2.866 on the hydropathy plot. A value that lies below the threshold value of zero demonstrates the active site is hydrophobic while a positive value demonstrates hydrophilic property.
References
Bernfeld, P., and Fishman, W.: (1953) Beta-Glucuronidase. I. Purification of Calf Spleen Beta-Glucuronidase , J. Biol. Chem. 202, p.757.
Bernfeld, P., and Fishman, W: (1950) Isolation of Beta-Glucuronidase of Calf Spleen, Arch. Biochem. Biophys. 27, p.475.
Buehler, H., Katzman, P., and Doisy, E.: (1951) Studies on beta-Glucuronidase from E. coli , Proc. Natl. Acad. Sci. USA 76, p.672.
Christner, J., Nand, S., and Nhatre, N.: (1970) The Reversible Inhibition of beta-Glucuronidase by Organic Peroxides, Biochem. Biophys. Res. Commun. 38, 1098.
Contractor, S., and Shane, B.:(1972) Purification and Characterization of Lysosomal beta-Glucuronidase from Human Placenta, Biochem. J. 128, p.11.
Dean, R.: (1974) Rabbit beta-Glucuronidase. Purification and Properties, and the Existence of Multiple Forms, Biochem. J. 138, p.395.
Glaser, J., Roozen, K., Brot, F., and Sly, W.: (1975) Multiple Isoelectric and Recognition Forms of Human beta-Glucuronidase Activity, Arch Biochem Biophys 100, p.536.
Oshima A, Kyle JW, Miller RD, Hoffmann JW, Powell PP, Grubb JH, Sly WS, Tropak M, Guise KS, Gravel RA (1987). Cloning, sequencing, and expression of cDNA for human beta-glucuronidase. Proc Natl Acad Sci U S A 84 (3): pp.685–9.