Introduction
The purpose of qualitative identification of microorganisms is to determine their biological species: to answer the question of what a particular specimen is. The identification task refers to classification studies, the outcome of which is to determine the nature of the organism. The practical value of such a task can be explained at several levels.
In terms of clinical implications, determining the nature of the bacterium provides an understanding of the nature of the pathogen and the appropriate measures that can be taken to eliminate the threat to health and life. In terms of laboratory implications, investigations help assess the contamination of samples, the success of their isolation and the assays performed. Such investigations also have forensic implications: the detection and further identification of microorganisms can be used for search efforts, suspect identification, and the establishment of evidence. In other words, the identification of unknown bacteria has many applied implications.
The primary tactic in an identification study is utilizing tests whose results consistently reduce potential conclusions. At the initial stage, there are usually no options for which bacterial species is present. Through successive microbiologic tests, it is possible to systematically reduce the outcomes and eventually arrive at one specific conclusion about the nature of the bacterium. Each of the tests is aimed at testing separate properties of the microorganism, which has the added advantage of identifying the enzymatic and biochemical features of the sample.
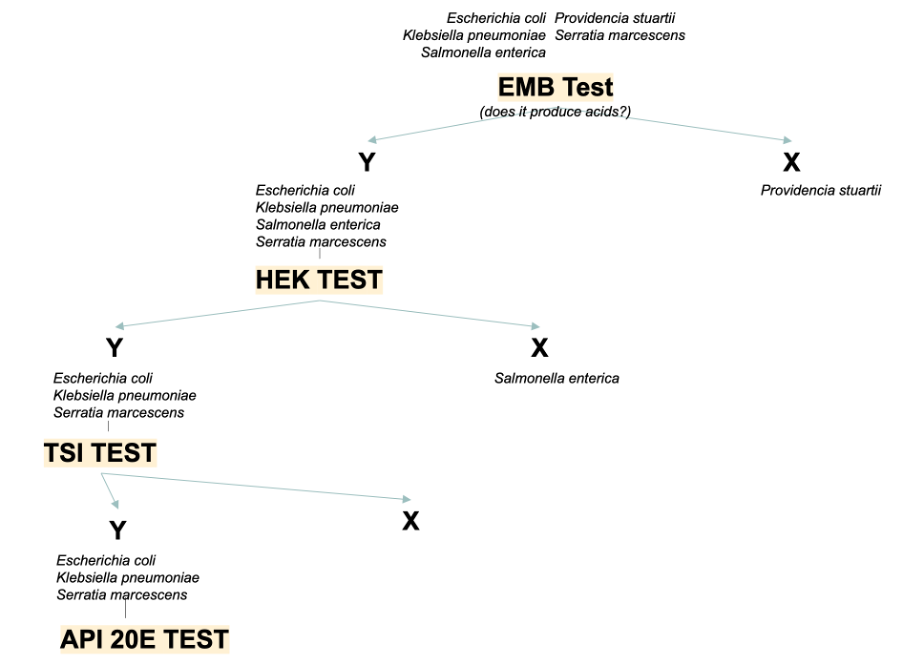
The present work aimed to identify an unknown bacterial species using a dichotomous key. The sample was given for microbiological tests, each of which allowed us to determine the response of the unknown microorganism to the proposed stimulus. Four tests were performed: the EMB, HEK, TSI slant, and API 20 E strip. First, the EMB test is used to determine the ability of Gram-negative bacteria to produce acid because the bromothymol blue indicator contained in the compound stains in the presence of excess protons.
Second, the HEK test is used to determine the enzymatic capacity of the microorganism to digest sucrose and urea. Third, TSI also allows the ability to determine sucrose digestion capacity because the citrate in the compound inhibits the growth of bacteria incapable of fermentation. Fourth, the API 20 E test is comprehensive: a tablet with different substrates allows multiple assessments of a sample’s ability to ferment several stimuli simultaneously.
Dichotomous Key
Figures/Tables
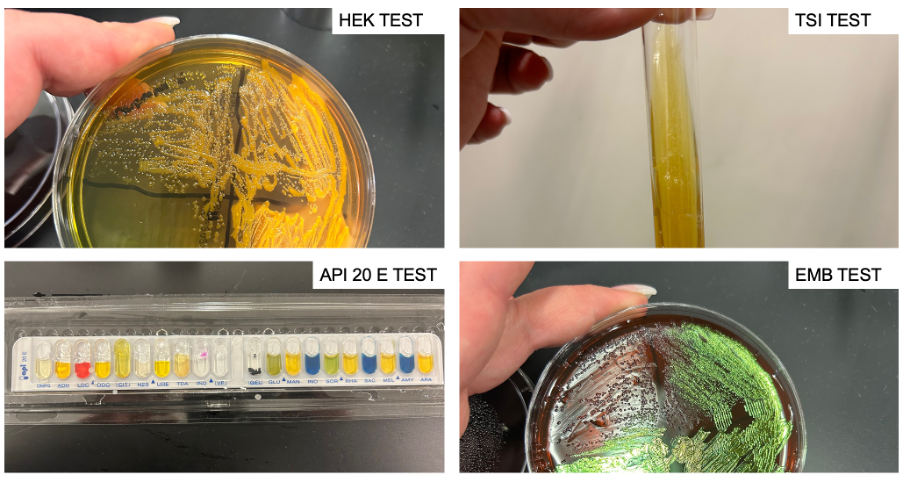
The results of the screenings performed using the dichotomous key (Figure 1) are shown in Table 1. Figure 2 shows photographs demonstrating the results of the microbiological tests performed.
Table 1. Results of the microbiological tests performed.
Conclusion
This work aimed to establish the nature of the unknown bacterium by performing a series of microbiological assays, as shown in a dichotomous key (Figure 1). Escherichia coli, Klebsiella pneumoniae, Salmonella enterica, Providencia stuartii, Serratia marcescens were used as candidates for the role of unknown sample. The sample showed a yellow bar across the surface, indicating the ability to digest sugars but no hydrogen sulfide formation): this left only Escherichia coli and Klebsiella pneumoniae on the list. In the EMB test, the sample showed green coliform bands, indicating the ability to ferment lactose; the list of candidates did not change. In the HEK test, the sample showed yellow staining, indicating the ability to synthesize acids; the list of candidates did not change.
Finally, in the multiple API 20E test, the list of positive and negative results of all 20 assays is shown in Table 1. From this data, it was evident that the sample contained ß-galactosidase, lactate dehydrogenase, capable of cleaving indole, capable of digesting glucose, mannitol, sorbic acid, maltose, rhamnose, and arabinose. After all the results, the unknown specimen sought was identified as Escherichia coli.
There were limitations to this study. First, EMB, HEK, and TSI slant results often overlapped, creating questions regarding which test to prioritize for a key. Having a standardized mechanism would have eliminated this limitation. Second, not all the assays within API 20 E provided noticeable results, which could also affect the quality of the resulting outcome. Addressing this limitation could have been done through repetition with a larger sample to decide whether to pass the test.