Introduction
Cell adhesion occurs on the substrate and requires cell adhesion molecules. Some of the essential cell adhesion molecules are cadherins, selectins, immunoglobulin and integrins (Humphries, Adam and Martin 3901). Types of cell junctions are anchoring junctions, occluding junctions, channel forming junctions and, signal relaying junction (Gumbiner 346). Inflammation illustrates cell adhesion. The cell adhesion molecules guide leukocytes to the injured area.
Failure of cell adhesion may create diseases, allow the cell to travel through the body, and metastasis (Gumbiner 347). Some of diseases that occur due to failure of cell adhesion are Leukocyte Adhesion Deficiency (LAD), Lesch-Nyhan syndrome (LNS), and Maple Syrup Urine Disease (Gumbiner 347).
Numerous cell adhesion assays employ microscopic observation and labelling of cells with a fluorescent dye for quantification (Spessotto, Katia, Pier, Eliana, Martina, and Roberto 227). It is essential to culture cells in the laboratory under sterile conditions in order to conduct in vitro studies (Hallab and Moses 211).
Cellular adhesion experiments are crucial in determining the success or failure of implanted material on a biomaterial surface (Gumbiner 348). Cell adhesion follows a recognizable sequence, hence making it easy to monitor through the use of fluorescent dye (Humphries, Adam and Martin 3902; Gallo, Masood, Jianjian, Reuben, Lei, Jennifer, Jianyun, and Randy 1690).
Cells should be attached to the substrate for efficiency in signal transduction. Signal transduction and elongation of cytoskeleton depends on the suitability of the material for cell adhesion. Attached cells undergo internal structural rearrangement and create internal tension (Hallab and Moses 215; Gumbiner 346). Actin get visualized using the protein phalloidin that binds strongly onto the actin filaments (Gumbiner 349). Adhered cells get stained with a fluorescent marker, phalloidin Rhodamine, in order to assess the extent of cell adhesion on surfaces, and examination of internal cell structure (Chazotte 6).
In order to monitor the mechanism of cellular adhesion, disruption of some stages in cellular adhesion is efficient through the application of Cytochalasin D. Cytochalasin D is an inhibitor of actin polymerization through binding to F-actin polymer. Actin filaments play a role in cell shape, motility and dynamics of the cell. Actin filaments are proteins, and have a function in mobility of the cell and its organelles. Cytochalasin D is a perfect inhibitor of mobility of cell organelles such as Golgi bodies.
Fixing the cell with 4% Paraformaldehyde helps in reducing the intensity of interaction with Cytochalasin. According to Glenn and Heptinstall (228), Cytochalasin D retards the incorporation of actin, myosin, and a 66 K protein into the cytoskeleton. F-actin contains more binding sites than G-actin that have affinity for cytochalasin D. Cytochalasin D inhibit actin filament elongation by binding to high-affinity sites located at the polymerization end of the filaments (Flanagan and S 835).
Tunicamycin is an antibiotic that inhibit enzymes involved in cell attachment and signal transduction such as UDP-HexNAc. Additionally, tunicamycin blocks N-glycans and inhibits cell cycle in the G1 phase (Ghoshal, Mythilypriya, Nadine, and Tohru 4). N-linked glycosylation is essential in cell-cell, and cell-extracellular matrix attachment.
Experiments which treat cultured cells with tunicamycin expresses suppressed P-selectin on the cell surface and in the cytoplasm. Glycosylation is critical for surface expression of P-selectin, and AKT signaling has a role in the tunicamycin-mediated inhibition of P-selectin (Jacob 606).
Results
The use of the fluorescent marker in the experiment aids in the examination of actin during cytoskeleton formation on the adhered cells. Cytochalasin D and Tunicamycin disrupt cytoskeletal assembly, and signal transduction. The presence of the actin cytoskeleton in cells is a crucial factor in the response of cells to the substrate.
Fixing and fluorescent labelling of cells helps in cytoskeleton visualization under Fluorescent Microscope. 4% paraformaldehyde is efficient in fixing adherent cells in cell culture. Rhodamine phalloidin has high affinity to the polymerized F-actin. The fluorescent signal helps in the visualization of the altered actin organization in the theramanox discs. The figures below show the results of the experiment where actin filaments absorb the red stain of Rhodamine phalloidin.
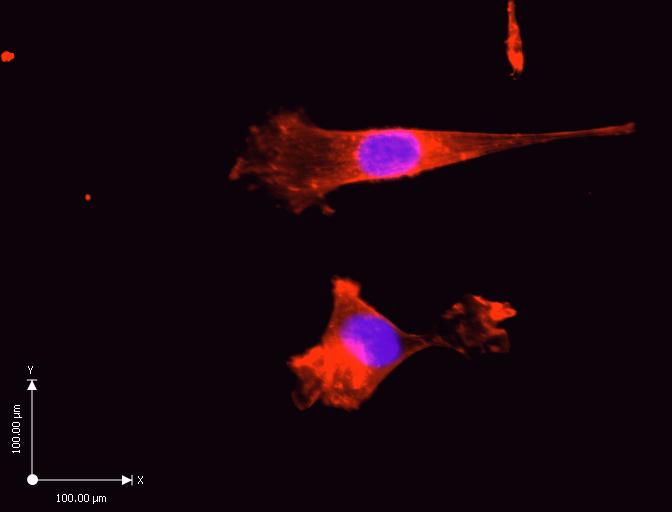
The results of the experiment in the control cells show an abundance of actin filaments between the cell, and the substrates. The reconstruction of cells shows a flat cell with stress fibers along the long axis, and homogenous coating with Rhodamine phalloidin. The measurements in the diagram give an estimate of the size of a single cell on the subrate.
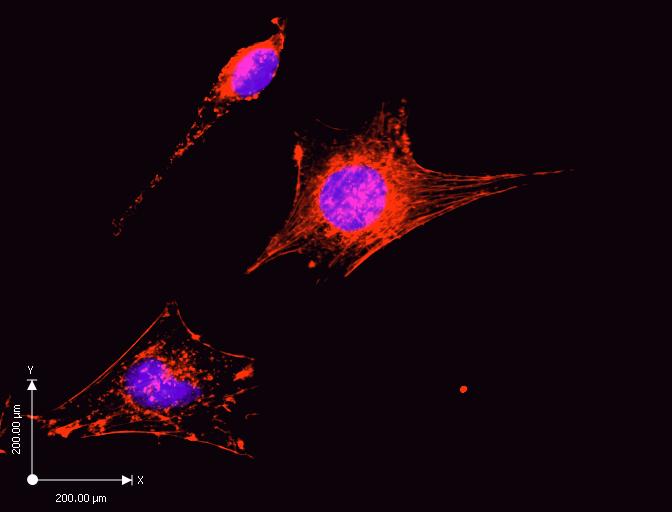
There is a minimal concentration of the actin filaments. The purple stain in the diagram represents the cell nucleus, whereas the red stain represents the actin filaments. Cytochalasin D treated cell has less dense actin filaments as indicted by the scattered red stains. Additionally, the measurements represent a high magnification of the cell for visualization. Cytochalasin D inhibited cell proliferation, and signal transduction because the cells are smaller than the control experiment.
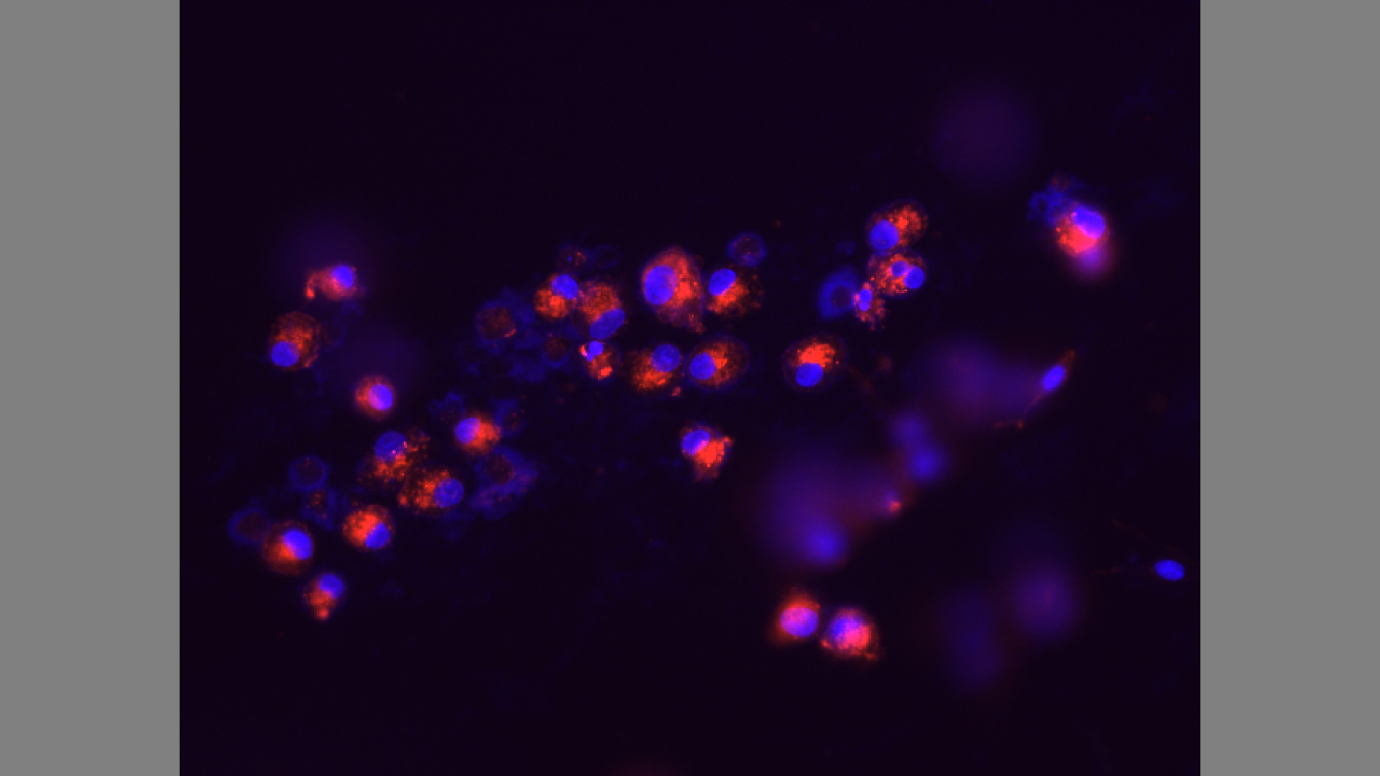
The cells are not attached to the extracellular matrix. Tunicamycin is toxic and makes the substrate unsuitable for cell attachment. Moreover, the actin filaments, as indicated by the red stain, are small. The experiment also shows signs of cell apoptosis from the smears of purple stain which represent the nucleus.
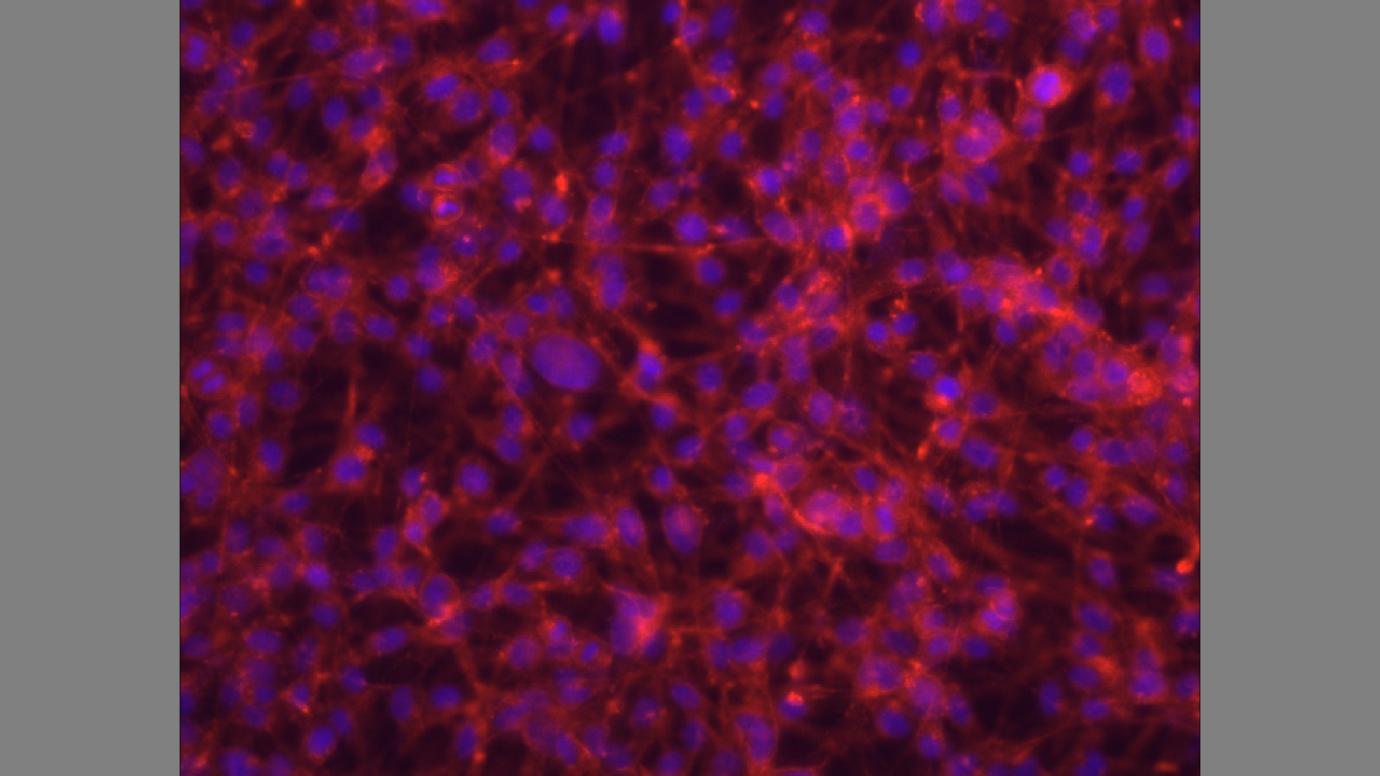
The cells express actin filaments, and P-selectin on the cell surface as indicated by the red stain on the substrate.
Discussion
Rhodamine phalloidin inhibits microfilament de-polymerization. Rhodamine characterizes, and identifies the constantly increasing number of microfilament proteins (Spessotto et al. 229). Figure 1 shows a control experiment of cultured cells that have not undergone treatment with Cytochalasin D, and Tunicamycin.
Rhodamine phalloidin has stained the actin cytoskeleton in cells that are abundant due to the suitability of the thermanox discs for cell adhesion, and also lack of toxins on the substrate (Gumbiner 353). The purple stain represents the cell nucleus, whereas the red stain represents the cytoskeleton fibers. Rhodamine phalloidin binds specifically to F-actin, and stains the actin stress fibers that are abundant in the control experiment (Gallo et al. 1691).
Figure 2 shows a snapshot of stained cells after addition of media containing Cytochalasin D. The snapshot shows the minimal concentration of actin filaments due to inhibition of association, and dissociation of subunits involved in actin polymerization. Additionally, the snapshot in Figure 2 shows shortened actin filaments. Cytocholasin D binds to affinity sites at the end of the filament blocking the elongation of actin filaments (Jacob 607).
Lack of elongation of actin filaments is an indication of the presence of high-affinity cytochalasin D binding sites in the cultured cells. The affinity of cytochalasin D for the binding sites corresponds to the effectiveness of the compounds inhibiting actin polymerization stimulated by F-actin. The experiment shows that the action of cytochalasin D is at the site of filament elongation. The results propose that the inhibitory effect of cytochalasin D on actin filament elongation receive mediation from the binding sites (Flanagan and S 837).
Figure 3 shows the effect of Tunicamycin on cultured cells. The cells are not attached to the extracellular matrix limiting signal transduction on cell-cell, and cell-extracellular matrix attachment. The actin filaments do not elongate leading to lack of cytoskeletal assembly. Tunicamycin inhibits glycosylation on P-selectin, and adhesion of cells on the thermanox discs (Jacob 609).
Tunicamycin does not affect mRNA expression leading into cell multiplication. It is evident that the Tunicamycin inhibits production of N-glycans, and modulated P-selectin expression from the experiment. Glycosylation is crucial for the cell surface expression of P-selectin in cultured cells. The results of the wells treated with tunicamycin show that the cells did not undergo glycosylation leading to negative results on cytoskeletal assembly (Ghoshal et al. 10; Gumbiner 354).
Figure 4 shows cytoskeletal assembly of cultured cells. The cells express P-selectin on the cell surface, and efficiency in glycosylation. The results show the adhesion mechanism of F-actin that generates tissue architecture. Cells adhere and send signals which control the transfer of information between cells (Gumbiner 355). The transmembrane glycoproteins mediate binding interactions at the substrate determining the cell-cell, and cell-substrate recognition (Hallab and Moses 220).
The results in figure 4 show the predicted cell behaviour, and cell responses which include spreading, migration, proliferation, differentiation, orientation, and apoptosis. The ability of cells to divide, proliferate, and migrate require an intact actin cytoskeleton that is visible through staining of actin filaments (Gallo et al. 1697). The actin filaments in figure 4 show a force-bearing structure that generates intracellular force to thermanox discs (Ochsner, Marcus, Viola and Michael 3).
Works Cited
Chazotte, Brad. “Labeling cytoskeletal F-actin with rhodamine phalloidin or fluorescein phalloidin for imaging.” Cold Spring Harbor Protocols 2010.5 (2010): 1-15. Print.
Flanagan, Michael D., and S. Lin. “Cytochalasins block actin filament elongation by binding to high affinity sites associated with F-actin.” Journal of Biological Chemistry 255.3 (1980): 835-838. Print.
Gallo, Richard M., Masood A. Khan, Jianjian Shi, Reuben Kapur, Lei Wei, Jennifer C. Bailey, Jianyun Liu, and Randy R. Brutkiewicz. “Regulation of the actin cytoskeleton by rho kinase controls antigen presentation by CD1d.” The Journal of Immunology 189.4 (2012): 1689-1698. Print.
Ghoshal, Pushpankur, Mythilypriya Rajendran, Nadine Odo, and Tohru Ikuta. “Glycosylation Inhibitors Efficiently Inhibit P-Selectin-Mediated Cell Adhesion to Endothelial Cells.” PloS one 9.6 (2014): 1-13. Print.
Glenn, JA May H. Ratan JR, and W. Losche P. Spangenberg S. Heptinstall. “GPIIb-IIIa antagonists cause rapid disaggregation of platelets pre-treated with cytochalasin D. Evidence that the stability of platelet aggregates depends on normal cytoskeletal assembly.” Platelets 9. 3-4 (2009): 227-232. Print.
Gumbiner, Barry M. “Cell adhesion: the molecular basis of tissue architecture and morphogenesis.” Cell 84.3 (1996): 345-357. Print.
Hallab, N. J., K. J. Bundy, K. O’connor, R. Clark, and R. L. Moses. “Cell adhesion to biomaterials: correlations between surface charge, surface roughness, adsorbed protein, and cell morphology.” Journal of long-term effects of medical implants 5.3 (1994): 209-231. Print.
Humphries, Jonathan D., Adam Byron, and Martin J. Humphries. “Integrin ligands at a glance.” Journal of cell science 119.19 (2006): 3901-3903. Print.
Jacob, Gary S. “Glycosylation inhibitors in biology and medicine.” Current opinion in structural biology 5.5 (1995): 605-611. Print.
Ochsner, Mirjam, Marcus Textor, Viola Vogel, and Michael L. Smith. “Dimensionality controls cytoskeleton assembly and metabolism of fibroblast cells in response to rigidity and shape.” PloS one 5.3 (2010): 1-12. Print.
Spessotto, Paola, Katia Lacrima, Pier Andrea Nicolosi, Eliana Pivetta, Martina Scapolan, and Roberto Perris. “Fluorescence-based assays for in vitro analysis of cell adhesion and migration.” Extracellular Matrix Protocols. Humana Press, 2009. 221-250. Print.