Introduction
Pectate lyase is an enzyme involved in the catabolism of pectate in cell walls of plants. Dubey et al. (2016) describe pectate as de-esterified pectin forming the main component of carbohydrates in the cell wall. Classification indicates that pectate lyase falls in the family of lyases, which cleave oxygen-carbon bonds in polysaccharides. Plant pathogens, such as yeast and bacteria, as well as symbiotic bacteria in the gut of ruminants, use pectate lyase in degrading pectate and deriving energy they need.
Hugouvieux-Cotte-Pattat et al. (2014) explain that pectate lyase applies the mechanism of β-elimination in degrading pectate to short polysaccharides comprising of 4-deoxy-α-D-galact-4-enuronosyl groups. In plant pathogenesis, pectate lyases contribute to the degradation of polysaccharides into oligosaccharides for other enzymes to continue with the degradation. Therefore, the understanding of structure and functions of pectate lyases plays a central role in their applications in industrial uses because they help in the catabolism of polysaccharides in cell walls, such as pectin and pectate. The emergence of molecular cloning has enhanced the application of pectate lyases in industrial processes of manufacturing natural fibres and fruit juices.
Pectate lyases are highly diverse because they vary according to their sources, structure and functions. Pectate lyase 1 (Pel1) is one of the established pectate lyases that has been created as a recombinant protein. The size of Pel1 gene is 1080bp that codes for an enzyme with 360 amino acid sequences. Cloning of this gene into an expression vector allows expression of its protein. Gay et al. (2014) recommend the use of expression vectors that are small and easy to clone, isolate and purify.
Therefore, the objectives of the series of experiments performed were to extract genomic DNA from Northumbrium northumbria, amplify Pel1 gene, clone it into the expression vector (pET-28a) and transform BL21 cells. Subsequently, the experiment aimed to express, purify, crystallise and characterise the recombinant Pel1. The overall objective of these experiments was to produce a recombinant Pel1 and determine its 3-D structure using x-crystallography.
Results
Extracted Genomic DNA
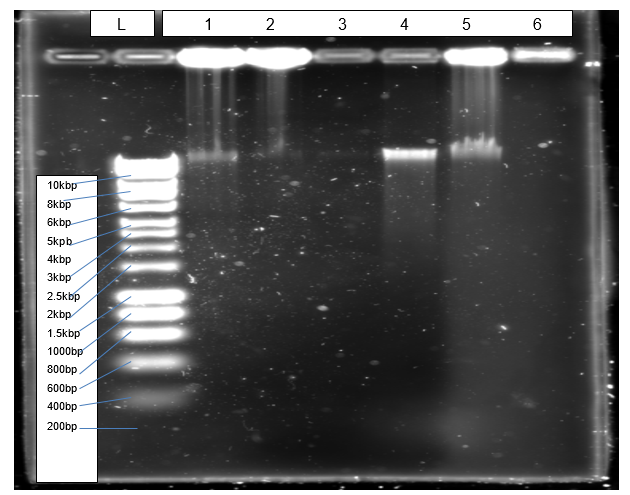
Figure 1 shows a clear band (4) of isolated genomic DNA from Northumbium northumbria.
PCR Product
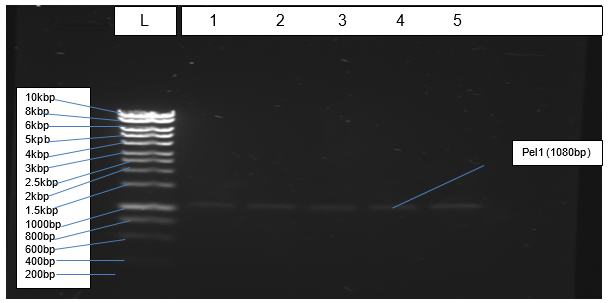
Figure 2 depicts results of PCR amplification of Pel1 gene resolving at the correct positions relative to the ladder.
Plasmid Purification
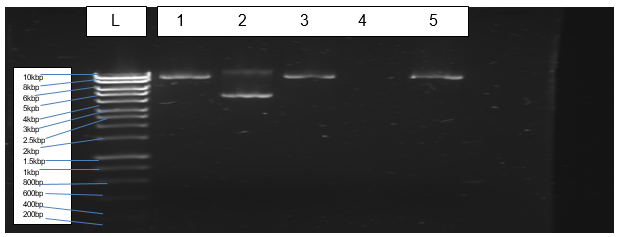
Figure 3 indicates bands of transformed and purified plasmids of Escherichia coli (BL21).
Concentration of Pel1 (ug/ul)
Absorbance = 0.055
Molar Absorptivity Coefficient (Ԑ) = 84800
Concentration (C)
Length (L) = 1
Relative molecular mass = 43203
A = ԐCL
Therefore, C = 0.055/84800L/mol*cm*1cm = 6.486⨉10-7 mol/L
Times dilution factor (10) = 6.486⨉10-6 mol/L
ug/ul = 6.486⨉10-6 mol/L*43203 = 0.28 g/L = 0.28ug/ul.
SDS-PAGE
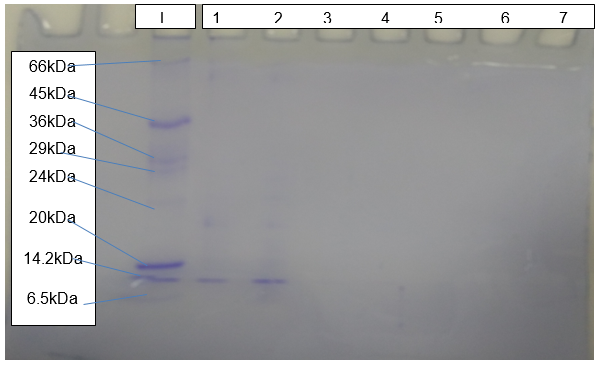
The outcome of SDS-PAGE (Figure 5) shows clear bands of protein resolving at about 15kDa relative to the standard molecular marker.
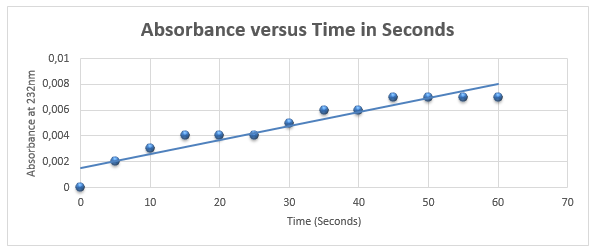
Activity Rate of Pel1
Figure 5 demonstrates that activity of Pel1 increases with time with the rate of 1.1 ⨉10-4 nm/s.
Activity = 103*1.11*10-4/43203*3ul = 8.82*10-8 U/ml
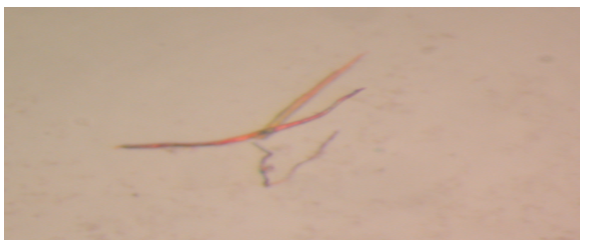
Crystallised Protein
Figure 6 is an image of plate-shaped crystals of Pel1 obtained from the expression of the gene in Escherichia coli.
Discussion
The objective of isolating genomic DNA from Northumbrium northumbria was achieved successful due to the formation of a clear band at the expected genomic size that is more than 10kbp (Figure 1). According to Mulcahy et al. (2016), a sharp band without smears indicates that an isolated genomic DNA is not also pure but also quality. The objective of amplification was achieved effectively because PCR generated the expected product of 1080bp, which resolved correctly in agarose gel (Figure 2).
The cloning of PCR products into the expression vector (pET-28a) was performed successfully. Restriction digests by enzymes (Xho I and Nde I) enabled ligation of Pel1 gene into the expression vector. Transformation of the E. coli (BL21) with the plasmid cloned with Pel1 occurred effectively. However, purification of plasmids did not materialise due to the loss of sample during purification.
Despite the occurrence of a human error during purification of plasmids, the expression of Pel1 was fruitful. Gay et al. (2014) hold that successful cloning and expression of the gene of interest is dependent on the size of an insert, the nature of a selectable marker and the presence of control elements. Based on the Beer-Lambert’s law, the molar absorptivity coefficient of 84800 L/mol.com and relative molecular mass of 43203, the concentration of Pel1 is 0.28ug/ul. Robinson (2015) explains that the determination of the concentration of enzyme forms the basis of calculating its activity. The SDS-PAGE reveals that the size of the expressed protein is about 15kD and crystal analysis shows that the size is 4kD.
In contrast, the expected theoretical size of Pel1 is approximately 35kDa. Guan et al. (2015) assert that systemic errors and modification of amino acid sequences explain differences in theoretical and experimental molecular weights of proteins. The activity rate of Pel1 is 8.82*10-8 Unit/ml, which is very low when compared to literature value of between 5.47 and 43.53 Unit/ml (Muslim et al., 2015). Eventually, crystallisation of Pel1 led to the successful production of plate-shaped crystals.
References
Dubey AK, Yadav S, Kumar M, Anand G and Yadav D (2016) Molecular biology of microbial pectate lyase: a review. British Biotechnology Journal 13, 1-26.
Gay G, Wagner DT, Keatinge-Clay AT and Gay DC (2014) Rapid modification of the pET-28 expression vector for ligation independent cloning using homologous recombination in Saccharomyces cerevisiae. Plasmid 76, 66-71.
Guan Y, Zhu Q, Huang D, Zhao S, Lo LJ and Peng J (2015) An equation to estimate the difference between theoretically predicted and SDS PAGE-displayed molecular weights for an acidic peptide. Scientific Reports 5, 1-11.
Hugouvieux-Cotte-Pattat N, Condemine G and Shevchik VE (2014) Bacterial pectate lyases, structural and functional diversity. Environmental Microbiology Reports 6, 427-440.
Mulcahy DG, Macdonald KS, Brady SG, Meyer C, Barker KB and Coddington J (2016) Greater than X kb: a quantitative assessment of preservation conditions on genomic DNA quality and a proposed standard for genome-quality. DNAPeerJ 4, 1-12.
Muslim SN, Al-Kademy IMS, Mahammed AN, Musafer HK and Muslim, SN (2015) Detection of the optimal conditions for pectate lyase productivity and activity by Erwinia chrysanthemi. Journal of Medical and Bioengineering 4, 184-191.
Robinson PK (2015) Enzymes: principles and biotechnological applications. Essays in Biochemistry 59, 1-41.