Introduction
An Overview of Genetic Engineering
Genetic engineering is one of the most fascinating fields of science that has changed different sectors of the economy. It involves a manipulation of the genetic makeup of an organism to produce novel or improved forms of the organism. Many scientists, especially in the medical, environmental, and agricultural sectors, have applied genetic engineering in solving real-life problems (Slama & Ziada 2016).
In environmental science, genetic engineering is applied in bioremediation where transgenic microbes that can degrade biological substances are used in waste management. In the medical field, recombinant DNA technology has been used to correct hereditary defects and in the development of vaccines. Transformation of bacterial cells, which is one of the approaches used in genetic engineering, involves the transfer of genetic material from one bacterium to another using a plasmid vector. The newly transformed bacterial cells can produce the desired products. The modified cells can be amplified (grown on a large scale) to produce sufficient quantities of the product.
Escherichia coli (E. coli)
E. coli is a gram-negative bacterium able to adapt to most environments (Rosano & Ceccarelli 2014). The E. coli genome is simple. Additionally, the organism has a haploid state throughout its lifetime. Therefore, mutations that normally happen during mitosis do not occur in this species. Under ideal conditions of bacterial growth, E. coli grows fast at the rate of one generation every 20 minutes, which translates to a fast production of the intended product. E. coli can also host foreign genetic material. Consequently, the preparation of competent cells before transformation with foreign DNA is easier when using this species than any other organism (Rosano & Ceccarelli 2014).
Green Fluorescent Protein (GFP)
Green fluorescent protein (GFP) is a protein isolated from jellyfish (Aequorea Victoria species). The protein is 26.9 kDa with 238 residues of amino acids. It exhibits bright green fluorescence when excited with UV light (Wright & Newman 2013). GFP is an excellent molecule for determining the expression of desired proteins in transformed cells.
The pGLO Plasmid in Transformation
The pGLO plasmid contains several genes with the notable ones being reporter genes (GFP and antibiotic resistance). Restriction sites for nucleases are present in the plasmid, which has a bidirectional promoter for the protein that metabolizes arabinose sugar and the gene that encodes GFP. An origin of replication (Ori) allows the plasmid to replicate autonomously and independently of the bacterial DNA. The plasmid contains a β-lactamase gene (bla) that can destroy β-lactam antibiotics (Ampicillin). The araC gene codes for an enzyme (protein) that metabolizes arabinose.
Regulation of Arabinose Operon in pGLO Plasmid
The regulation of the arabinose operon is a complex process involving various mechanisms. The original arabinose operon controls the expression of different proteins that are encoded by three structural genes (araB, araA, araD or BAD) (Rosano & Ceccarelli 2014). Apart from these genes, another gene (araC) codes for the synthesis of a protein that directs how BAD is expressed (Lee et al. 2017).
The araC gene is also a repressor of its own synthesis through the binding of its product to itsoperator and preventingthe transcription of thegene. BAD is expressed when araC protein and arabinose sugar are present. The attachment of arabinose to araC gene causes conformational changes in the regulatory region (activator) of the BAD genes. Subsequently, RNA polymerase is recruited to a precise location (promoter region) in the upstream of the BAD gene leading to the transcription of the three genes (Wright & Newman 2013). The negative regulation of araC expression leads to the activation of the BAD genes and vice versa.
The purpose of this experiment was to provide practical skills in the genetic manipulation of an organism through transformation. This report explains the transformation of the bacteria (E. coli cells) with the pGLO plasmid.
Materials and Methods
The E. coli cells used in this experiment were retrieved from an earlier prepared culture. Isolation of colonies was done at standard temperature and pressure. Two Eppendorf tubes were labelled + pGLO and –pGLO and placed on a foam rack. 250 µL of calcium chloride solution (transformation solution) was added to each of the tubes, which were then placed on ice for 5 minutes. A single E. coli colony was picked up using a sterile loop and placed in the bottom of the two tubes containing the transformation solution. The loop was spun severally using the thumb and index finger until the entire colony was dispersed into the transformation solution. Plasmid DNA solution was then introduced into the suspension of bacterial cells in the +pGLO tube only using another sterile loop. The two tubes were incubated on ice for 10 minutes.
Four agar plates were labelled. The first plate was labelled “+pGLO” and was used to hold LB media containing Ampicillin (LB/Amp plate). The second plate was labelled “+pGLO” and represented the LB/Amp/Ara plate. The third and fourth plates were labelled “-pGLO” and represented the LB/amp and LB plates for the negative controls. The +pGLO and -pGLO tubes were transferred into a water bath maintained at 42 °C for exactly 50 seconds.
The purpose of this step was to open the cell membranes briefly and allow the plasmid to get into the cell. The tubes were then placed on ice for 2 minutes. A germfree pipette tip was used to transfer 250 µL of LB nutrient broth into each of the tubes followed by tapping to mix the contents of the tubes, which were then incubated at 37 °C for 30 minutes in a shaking incubator.
The bacterial cells were then resuspended by tapping. About 100 µL of the transformed cells were added to the two plates labelled LB/Amp and LB/Amp/Ara. A similar quantity of untransformed cells was added to the “-pGLO” plates (LB/amp and LB plates). The surfaces of the plates were then streaked evenly, sealed by taping and incubated at 37 °C overnight. After that, the plates were transferred to the cold room for storage until further analysis.
Results
Figure 1 (A) shows the LB plate with the pGLO plasmid. There was a dense growth of bacteria on this plate. Figure 1 (B) shows the LB/AMP plate without the pGLO plasmid. No bacteria grew on this plate after 24 hours of incubation at 37 oC. These results show that the untransformed bacterial cells were unable to grow on media containing Ampicillin.
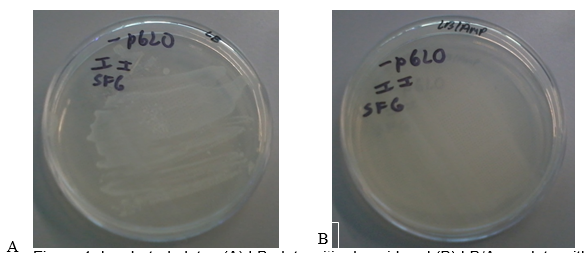
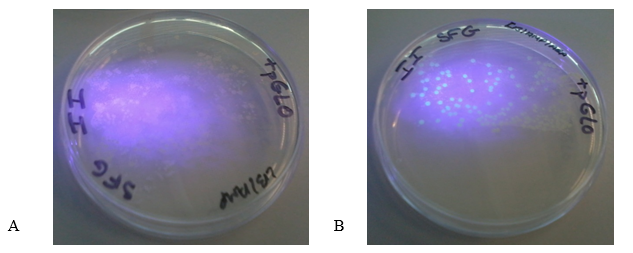
Figure 2 (A) shows the LB/Amp plate with the pGLO plasmid. The colonies appeared white under UV light. Figure 2 (B) shows LB/Amp/Ara plate with the plasmid. The colonies appeared green underneath the ultraviolet light thus indicating the expression of GFP. These findings show that GFP is only expressed when transformed bacteria grow on media containing arabinose.
Discussion
The goal of the experiment was to transform E. coli cells using the pGLO plasmid, which contains two distinct reporter genes (the Ampicillin resistance genes and GFP genes). Therefore, the success of the experiment was dependent on three essential factors: the use of a β-lactam antibiotic (Ampicillin) for bacterial selection, the presence of arabinose in the medium, and expression of the GFP protein.
The bla gene in the pGLO plasmid is responsible for antibiotic resistance through the production of β-lactamase that degrades Ampicillin (Akbari & Akbari 2017). Therefore, all transformed cells survive in the presence of Ampicillin. This phenomenon explains why the transformed bacteria in Figure 1 A grew into a dense colony because the plasmid conferred antibiotic resistance, thus enabling them to thrive in media containing Ampicillin. On the other hand, the bacteria in Figure 1B did not grow on media containing Ampicillin because they did not contain the pGLO plasmid (they lacked antibiotic resistance).
In successfully transformed cells, GFP is expressed when arabinose is contained in the growth medium, which allows the detection of the protein under UV light (Akbari & Akbari 2017). The bacteria in plates A and B in Figure 2 were transformed successfully and contained the pGLO plasmid. The presence of arabinose in the media triggers the expression of GFP, whereas the absence of arabinose in the growth medium blocks the transcription of the arabinose metabolizing gene (Muralidhar et al. 2016).
The colonies in the plate B fluoresced green under UV light because of the expression of high levels of GFP in the presence of arabinose (Akbari & Akbari 2017). The presence of arabinose in the medium caused araC protein to change its conformation thus enabling RNA polymerase to bind the promoter and transcribe the GFP. However, the cells in the plate B appeared white upon illumination with UV light because araC blocked the expression of GFP due to the absence of arabinose in the medium.
The findings of this experiment corroborate the findings of previous studies, for example, an investigation conducted by Bennett, Chau and Ma (2015) to find an effective way of detecting gene expression in transformed E. coli cells. An interesting observation, however, is that GFP was expressed within 40 minutes following the induction of transformed bacteria with arabinose. The amount of GFP is proportional to the amount of green fluorescence emitted following UV illumination.
Also, the level of GFP expression increases with the length of incubation time. Bennett, Chau, and Ma (2015) reported that very little fluorescence was observed in the initial 30 minutes after induction of transformed E. coli with arabinose. However, the intensity of the fluorescence increased gradually reaching a threshold at the 60-minute mark.
In conclusion, the pGLO plasmid was used to transform E. coli cells successfully indicating that the transformation protocol was effective. Transformed cells survived on media containing Ampicillin thus showing the efficacy of selective media in identifying transformed cells. The expression of GFP only occurred when arabinose was incorporated in the growth medium thereby demonstrating the importance of arabinose in the expression of the araC gene in the plasmid.
References
Akbari, DL & Akbari, LF 2017, ‘Detection of GFP expression and colonization of wheat by two Endophytic bacteria tagged with GFP gene’, International Journal of Pure and Applied Biosciences, vol. 5, no. 1, pp. 844-848.
Bennett, D, Chau, CH, & Ma, R 2015, ‘Investigating flow cytometry as a potential method for real-time analysis of gene expression following Escherichia coli transformation’, Journal of Experimental Microbiology and Immunology, vol. 15, pp. 1-6.
Lee, DJ, Sánchez-Morán, E, Bryant, JA, Sellars, LE, Sánchez-Romero, MA & Busby, SJ 2017, ‘Development of a new fluorescent reporter: operator system: location of AraC regulated genes in Escherichia coli K-12’, BMC Microbiology, vol. 17, no. 1, p. 170.
Muralidhar, Y, Sravanthi, M, Prasad, TNVKV, Nissipriya, M, Reddy, PS, Neelima, TS, Reddy, GD, Adilaxmamma, K, Kumar, PA & Krishna, TG 2016, ‘First report on rapid screening of nanomaterial-based antimicrobial agents against β-lactamase resistance using pGLO plasmid transformed Escherichia coli HB 101 K-12’, Applied Nanoscience, vol. 6, no. 6, pp. 941-949.
Rosano, GL & Ceccarelli, EA 2014, ‘Recombinant protein expression in Escherichia coli: advances and challenges,’ Frontiers in Microbiology, vol. 5, p. 172.
Slama, RA & Ziada, AS 2016, ‘Initial stages of construction of a plasmid to study the kinetics of gene expression at a single cell level following uptake of DNA into Escherichia coli’, Journal of Experimental Microbiology and Immunology (JEMI), vol. 20, pp. 86-91.
Wright, LK & Newman, DL, 2013, ‘Using PCR to target misconceptions about gene expression’, Journal of Microbiology & Biology Education: JMBE, vol. 14, no. 1, pp. 93-100.