The objective of the experiments was to down-regulate expression of two vital genes, unc-22 and lsy-2, responsible for respectively, muscular integrity and chemotactic perception in soil nematode, Caenorhabditis elegans. For this purpose, RNAi technique was used, in which two plasmid vectors, pLT61 (anti-unc-22) and pX1P11 (anti-lys-2), were allowed to multiply in Escherichia coli strain HT155. The genes were allowed to express as dsRNA under the promoter control of T7 RNA polymerase. These bacterial cells were fed to C. elegans so that natural transformation of dsRNA into the somatic cells of the worm takes place. Subsequently, the knock-down or gene silencing effect of dsRNA was tested in the worm by way of their crippled movement (unc-22 under-expression) or failure of chemotactic movements (lys-2 under-expression). If these anti-sense RNA’s knock down the corresponding sense RNA’s, then against the control worms the above phenotypic changes will be discernable.
Introduction
RNAi is an effective technique to selectively knock-down the gene expression in number of eukaryotic organisms, including human and worms. Investigation on RNAi mechanism in C. elegans has direct relevance in human to decipher many homo- and orthrologous genes that may have same function to that of the worm (Grishok 5932-5939). The homologues genes of unc-22 and lys-2 in human are associated with diseases of muscular and neural developmental process. Unc-22 homologue, titin, a spring-like protein in sarcomere of muscle microfilaments, is responsible for proper positioning of actin and myosin filaments in sarcomere (Alberts et al. 963-964), and its deficiency leads to Down Syndrome. The RNAi plasmid vectors used are derived from pL4440, which has ampicillin resistance marker brought from pBR322 and a multiple restriction site for cloning between two T7 RNA polymerase promoters, which operates in presence IPTG under the lacO control. A transformable tetracycline resistant bacterial strain, HT115, was suitably designed to produce T7 RNA polymerase. The strain is also devoid of RNaseH, responsible for breakdown of dsRNA. Thus any gene cloned between T7 polymerase promoters will be expressed as dsRNA. In the strain of C. elegans (NL2099) used here, RNAi-antagonizing gene, rrf-3 (RNA-dependent RNA polymerase), has been deleted, so that RNAi mechanism is not interfered. The above RNAi vectors were used for unc-22 and lys-2 down-regulation.
Results
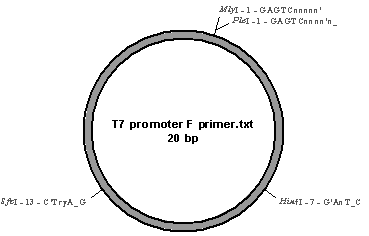
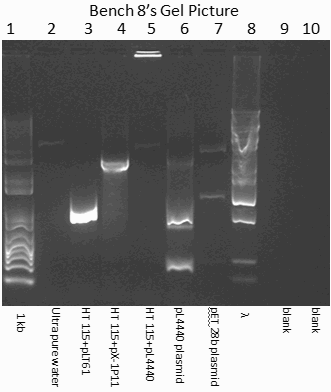
Apart from bacterial strains with RNAi vectors (HT115 (DE3)-pLT61 and HT115–pX-1p11), a strain harboring empty RNAi plasmid (HT115-pL4440) was chosen as control for identification of PCR products. Pure uncut plasmid DNA’s, pL4440 with two T7 RNA polymerase promoters, and pET28b with only one promoter, were also used as controls. As shown in Table 1 (D1.3), these DNA’s (lysates of bacteria and plasmid DNA’s) were amplified with T7 promoter-specific primer from either sides of the inserted RNAi clones. The PCR products thus produced were analyzed by 1% agarose gel containing SYBR green dye. As given in Table 2 (D3.1), 1+Kb and Lambda BstEII digest markers were included on either side. Ultra-pure water was loaded in one of the wells as negative DNA control. Upon completion of electrophoresis the regions of DNA fluoresced green/orange under UV. Figure 1 shows a pDRAW diagram of RNAi plasmid with T7 forward primer position. As shown in Fig. 2 (bench # 8), the results in lanes 1-8 were as follows: 1+Kb marker (ca. 15 bands; 100-12000 bp), ultra-pure water (no band), H115-pLT61 (one band), H115-pX1P11 (one band), H115-pL4440 (no band), pL4440 (two bands), pET28b (no band), Lambda BstEII digest (ca. 10 bands; 702-8453 bp). The relative positions of the bands were measured from the wells and by matching with the standard DNA ladders, approximate molecular weights of the PCR products were calculated (Table 1 (D1.3)) as follows: H115-pLT61 = 1,300 bp, H115-pX1P11 = 3,600 bp, and pL4440 = 300 + 900 bp. In the other gel only 1+Kb ladder could be resolved and Lambda ladder was disorganized. Most likely the PCR amplification did not yield sufficient DNA for visualization in this gel.
PCR questions:
- Here, only one T7 promoter forward primer was used. This is because the gene of our interest is cloned in between two T7 promoters of identical sequence. Hence during PCR amplification the same forward primer would act as reverse primer from the other side, and both the transcripts will be read in opposite direction (5’ ends) giving two complementary RNA strands, which would pair as dsRNA.
- As pL4440 is without any RNAi gene, only that portion of DNA which is flanked between the two T7 RNA polymerase promoters, mostly comprising the multiple restriction digestion sites would be amplified. In H115-pL4440 there was no band, suggesting that this piece of amplified DNA was too small to be seen in the gel. However, intact pL4440 amplified in two bands, which seem to be coming from some contaminating DNA present in this preparation.
- The size of amplification product of pLT61 is ca. 1,300 bp from this result and from records it was found to be ca. 1,000.
- We do not expect any amplified product from pET28b because it has only one T7 RNA polymerase promoter primer sequence. Thus the reverse amplification would not be possible even if any gene is cloned under this promoter control.
- The objective was to check whether the RNAi inserts in the vectors were still present in the given bacterial strains after growth. If the bacteria harbored only the RNAi-free vectors like pL4440, then we should have not seen any amplification, and this is what the result was. The apparent contamination of ca. 300 and 900 bp DNA’s of pure pL4440 preparation was not present in the lysate. As we found only one band of amplicons in H115-pLT61, H115-pX1P11, it is apparent that the test bacterium is capable of producing dsRNA from the two inserts, unc-22 and lys-2.
- The size of PCR product of RNAi lys-2 in HT115-p1P11 RNAi vector, upon bacterial lysis was deduced to be ca. 3,600 bp.
After confirming that the E. coli H115 harbored C. elegans unc-22 and lsy-2 genes in their plasmid vectors and one control strain was devoid of any such RNAi insert, the bacterial cells were fed in three batches to the worms. The worms fed with bacteria harboring pLT61, pX1P11 and pL4440 were analyzed for overall morphology and counts under a dissecting microscope. Table 3 (D3.2) presents the data on phenotypic characteristics of the three batches of worms. There was a minor increase in the worm body length in case of pLT61 and pX1P11-fed animals when compared with pL4440 fed controls.
Table D3.2 related questions
- In controls there were 50 large worms, and the number significantly reduced in case of unc-22 and lys-2 RNAi animals (n = 20-25).
- Yes, the worms still might possess the ability to lay eggs. According to literature, the RNAi effect can be seen in next generation (F1) and sometimes even further. The number of large (matured) hermaphrodite worms in all the three sets with apparent normal morphology must be having brood and can lay eggs.
The phenotypic characterization of C. elegans after RNAi was carried out by two procedures. Since unc-22 gene under-expression (pLT61) is associated with movement defects, such animals were compared with controls (pL4440) in normal NGA plates in terms of their circular twitched or S shaped rattling motion. The lys-2 gene under-expression affects the lys-6 gene transcription, which is associated with chemotactic response towards NaCl. Chemotaxis plates were set up with two spots, one of NaCl and another of water. Another set of plates had only two water spots. The worms fed on bacterium harboring pX-1P11 were transferred to NaCl (Na) + water (W) spotted plates and designated as set B. Control worms fed on bacterium with pL4440 were also placed similarly (set C1). The same control worms were also placed on plates having two water spots (set C2). As shown in Table 4 (D3.3) there was a clear difference in the movement characteristics between the worms with unc-22 RNAi and those without. In the RNAi worms the head twitched, and the worms started to move spirally and there was no rattling. Some of the worms also died. On the other hand, in controls the S shaped motion and rattling of the fully viable worms was seen.
There was some unusual pattern of chemotactic distribution found in the lys-2 RNAi and control worms with respect to movement towards NaCl spot and crowding in the region of high salt (Table 5 (D3.4)). In plate C1 which had the non-RNAi worms (controls), nearly 1.5 times more worms migrated towards salt than those towards water. In contrast, with lys-2 RNAi (plate B), where the movement towards salt should have been inhibited, the worms in fact migrated more towards salt (3-times to that of water). This difference does not explain retarded chemotactic response due to RNAi, because most of the worms (both plates) were still present in central spot and their migration pattern could not be ascertained beforehand. The control worms moved almost equally and randomly towards both water spots (plate C2). Table 6 (D3.5) is percentage representation of Table 5 (D3.4) data, and one can see that a high proportion of total population is still present in central spot. Table 7 shows a summary of the chemotaxis results from each bench in the section. Analysis of the results from all 8 batches of chemotaxis plates gave some unusual trends. With some exceptions like in bench #7, control worms in C2 plates distributed almost evenly between the two W spots. Control worms however showed a preferential migration towards Na spots in most of the C1 plates. Contrary to this, lys-2 RNAi worms showed a very discrete pattern as far as distribution between Na and W spots in B plates is concerned. Either the worms were evenly distributed (bench #1, 2, 6), or they exhibited preferential migration towards Na (bench #3, 4, 5, 7, 8). Another intriguing observation is that if starting population in central spot is high then most of the worms did not move from central spot and the motile ones preferred going to Na spot (e.g. bench #4 and 7).
Table 1 (D2.3): Expected Amplicon size
Table 2 (D3.1): Sample application in different lanes
Table 3 (D3.2): Visual analysis of C. elegans
Table 4 (D3.3): Analysis of C. elegans after unc-22 RNAi
Table 5 (D3.4): Chemotaxis count
Table 6 (D3.5): Percentage of worms
Table 7: Summary of Chemotaxis count results from each batch
Bench 1
Bench 2
Bench 3
Bench 4
Bench 5
Bench 6
Bench 7
Bench 8
Discussion: Addressing the five questions each in one paragraph
Using only one forward T7 polymerase promoter primer, the two RNAi vectors in bacterial lysates, H115-pLT61 and H115-pX1P11, were expected to generate one amplicon each, corresponding to the inserted unc-22 and lsy-2 genes expressed under control of T7 promoters. A small size PCR amplicon was also expected from RNAi empty H115-pL4440 corresponding to the multiple restriction cassette. But we did not expect any PCR amplification in case of pET28b as it had only one T7 promoter. The Lambda and 1Kb markers were expected to give ladders of designated molecular sizes. Upon feeding the worms with H115-pLT61 and H115-pX1P11, we expected phenotypic changes due to down-regulation of respectively, unc-22 and lsy-2 genes in the worms. The former is expected to adversely affect normal rattling motion and viability of the worms and the later would have retarded chemotaxis towards NaCl. Controls fed with H115-pL4440 were without any RNAi and were expected to exhibit S shaped motion and chemotactic behavior. Compared to controls, in both RNAi worms we expected retarded growth and less number of large adults, as the knocked-down genes were related to development process. Indeed the adults were fewer in number in the RNAi worms than in controls.
In gel, only one band each was detected corresponding to amplification products of unc-22 and lsy-2 in H115-pLT61 and H115-pX1P11 lysates. Both the markers resolved well and produced ladder of designated sizes. No band in case of pET was also as per expectation. There was some ambiguity between H115-pL4440 and pL4440 amplification results, while former did not amplify to yield a band, the later gave two bands. Most likely, the pL4440 preparation had low molecular weight DNA contamination. The RNAi worms’ size was more than controls, and this explains that there is no stunted growth due to gene knock-down, only the numbers reduced. The H115-pX1P11 RNAi worms’ chemotaxis behavior went quite against our expectations. There was no consistency between the benches. The justification is that only limited number of worms acquired the dsRNA through feeding H115-pX1P11 and thus in several experiments they moved equally well towards Na and W spots. The others in which RNAi activity did not take place behaved more like the controls and migrated to Na spot.
The chemotaxis plates presented an ambiguous result due to experimental errors (D3.4). The worms were randomly transferred to the plates without considering their size and number. May be the juveniles and small worms are only partially affected as they might not feed much bacterial cells and depend more on reserve food. There was also a crowding effect in central spot and probably this has restricted the worms to move away from this area.
The pL4440 preparation should be resolved on gel to verify whether the DNA contamination still exist, and if so, then a new batch should be used for PCR amplification. To evaluate the phenotypic properties of RNAi it is necessary that the controls and RNAi worms should be taken of identical age (size), preferably the larger ones, and their numbers should be nearly equal (ca. 50). This would avert the crowding effect in chemotaxis plate, and as the worms have spent their entire life stages feeding on bacteria, the RNAi phenotypic effect would be more pronounced. Only one set of D3.3 result is also not enough to arrive to any conclusion, even though the results are as per the expectation. Statistically, multiple experiments starting with equal number and identical age of worms would provide a clearer picture on abnormal phenotypic characteristic as the case may be (twitching, chemotaxis).
The down-regulation by dsRNA tested in this study was more symptomatic and based on phenotypic changes in the worm. It would be the next course of action to verify as to how much RNAi effect was found in the worm. For this, unc-22 and lsy-2 expression at transcript and protein level in control and RNAi worms should be evaluated. The total RNA and protein can be extracted from whole animals and can be subjected to Northern blot and immunoblot analyses using appropriate DNA probes specific to mRNA and antibodies for the gene products. This would give a clear idea on connection between RNAi and associated phenotypic changes at cellular level.
Report Questions on page 110 (only the answers):
- unc-22 codes for a sarcomere protein, twitchin. This protein anchors the Z-disc of muscle microfilaments with the thick myosin fibers. It has a spring like action and upon any stress activity it readily relaxes or contracts itself and adjusts the necessary position of actin and myosin fibers at regular order by a mechanism that involves protein phosphorylase/kinase action. In C. elegans whose sarcomeres are longer than vertebrates UNC-22 is also longer, and it serves as molecular ruler determining the overall length of each sarcomere. RNAi knock-down would affect the muscle development and the worms would move spirally rather than in S shape. Their viability is also expected to be affected.
- The two C. elegans gustatory neurons, left and right ASE are laterally similar but are asymmetric in function and some expression of chemosensory signaling mechanism is also different. A complex gene regulatory mechanism by microRNAs regulates the developmental process of neurons. LYS-2 is a novel transcriptional factor controlling the synthesis of this microRNA. Alterations in lys-2 switch off this microRNA synthesis and consequently this adversely affects the complete lateral neural system, particularly those nerves responsible for chemotaxis.
- This is the most suitable control in which RNAi dsRNA would not be synthesized. This control would rule out any feeding-related changes in the overall phenotypic characteristics of the worms other than RNAi. As alternate bacterial strain, BL21(DE3) is also appropriate, because it is naturally transformable and has T7 RNA polymerase gene cloned for expression of any vector that has T7 promoter primers. Using the two bacterial strains, feeding related difference in phenotypes can be nullified from RNAi related changes.
Bioinformatics Questions for Lab Report (only the answers):
- The other name of unc-22 protein product in C. elegans is twitchin, a giant intracellular protein homologous to mammalian protein called titin. UNC-22 is required in muscle for actomyosin contraction-relaxation cycle and for maintenance of normal muscle morphology. UNC-22 associates with myosin and is localized to A-bands. In vitro UNC-22 phosphorylates myosin light-chain peptides. Once knocked-down the myosin phosphorylation would stop and hyperphosphorylated bands would not take its position in juxtaposition with actin and thus muscle relaxation would be badly affected.
- lsy-2 encodes a novel C2H2 zinc-finger transcription factor that is most closely related to other nematode zinc-finger proteins. LSY-2 is required for the left-right asymmetry of the ASE neurons. As transcriptional factor for the lsy-6 microRNA, it controls the asymmetric expression of guanylyl cyclase chemoreceptors. Further, this microRNA affects expression of cog-1, required for full functional development of right side of nervous system in C. elegans. Its deficiency can affect the overall development of both right and left side of nervous system and overall chemo-perception would be collapsed.
- According to OMIN search, human protein with a similarity to unc-22 gene product is titin. Mutation in this gene in human is associated with muscular dystrophy, like hypertrophic cardiomyopathy due to hyper-affinity of titin to alpha actin, Tibial muscular dystrophy manifested by weakness and atrophy of anterior lower leg, and Edstrom myopathy owing to mutation in protein kinase domain. In human I band of titin makes spring-like connections between the thick filament and the Z line within the sarcomere. The A-band binds to the other side with myosin thick filament, regulating the filament length and assembly. There are two elements within I band region that correlate with sarcomere stiffness. Hence, two springs may act in a series giving versatility in sarcomere length and resting muscle tension in human.
- In C. elegans, rde-4 codes for dsRNA binding protein, which recognizes and then cleaves dsRNA to small interfering (si)RNA’s. In addition to making a complex with dsRNA, RDE-4 also binds to DCR-1 (RNase III related enzyme), DRH-1 (helicase), and RDE-1. Collectively, this Dicer complex cuts the dsRNA to siRNA, to which the RDE-1 remains bound until completion of RNAi event (Grishok 5933). Mutation in rde-4 restricts this assembly to form, and consequently siRNA synthesis is inhibited and RNAi activity is lost. C. elegans rde-4 has no human homologue, but a related RNAi gene, rde-1, has similarity to Eukaryotic translation initiation factor 2C-3 in human that has a function other than RNAi in protein synthesis.
Questions to be considered when analyzing effect of RNAi on your worms:
- Yes, the dsRNA is reported to survive up to F1 and hence RNAi related phenotype for unc-22 would appear in the next generation. Besides, a significant proportion of RNAi worms grew to large size and reached adulthood. As they are hermaphrodite, they can lay eggs and produce F1 progeny with the parental phenotype of twitching movements and reduced viability.
- Proteins make complex with mRNA and thereby protect the RNA from pairing with the corresponding siRNA, a step that is essential for RNAi. If the protein turnover is high means half life is short, then less protein will be accessed by the mRNA for binding, and therefore the cutting sites will be open to RNAi. A better down-regulation can be seen if such proteins are degraded faster than synthesized.
- The 100% normal worm did not move towards Na spot. This means within this population some proportion had no or retarded chemotaxis phenotype. While there was no lsy-2 RNAi in controls, some microRNA (lsy-6) expression must have been still affected by unknown mechanism. One likely reason could be transposon-mediated mutagenesis that is fairly common in C. elegans. Variation in relative migration in controls does not allow us to conduct ANOVA to figure out how significant the difference is within the controls. Naturally, comparison with the RNAi becomes even more incorrect. One method to counter this problem is to take population derived from single parthenogenic parent. Expectedly, the phenotype within the population would not be much different. The starting number of worms should also be the same to make a better statistical analysis of distribution within the control groups.
- In some of the tables, significant variation in relative distribution of worms between Na and W spots was found. This problem was encountered more in case of RNAi worms (Plates B). Those batches in which the tables showed that the worms preferentially moved towards Na spots (bench #3, 4, 5, 7, 8) or were crowded in the central spots should be disregarded, as RNAi was apparently not successful in these batches and the worms behaved more like the control ones.
References
- Alberts, Bruce, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, and Peter Walter. Molecular Biology of the Cell (4th Ed.). New York: Garland Science Taylor & Francis Group, 2002.
- Grishok, Alla. “RNAi mechanisms in Caenorhabditis elegans.” FEBS Letters 579 (2005): 5932-5939.
- OMIN (Online Mendelian Inheritance in Man). Johns Hopkins University. 2008