Introduction
The non-structural protein 5 (NS5) is considered to be the largest strain, which is coded by the Zika Virus RNA. According to Elshahawi et al. (2019), the product has about 904 amino-acids along its entire length. It is comprised of two domains, which complement each other’s function. In particular, it has an RNA-dependent polymerase (RdRp) domain, which is located at its C-terminal and is joined to a methyltransferase (MTase) at its N-terminal through a linker. The NS5 strain is a critical element in the replication of Zika Virus. Moreover, it is also important in the survival and evasion of immunity of the system it enters, in addition to other functions. According to Upadhyay et al. (2017), ZIKV NS5 protein is similar in several structures to the Japanese encephalitis virus (JEV). This paper hypothesizes that the Zika Virus mutates, thus varies from different geographical locations and time, and NS5 ensures its survival and infectivity.
Methodology
Zika envelope genome. Every Zika virus genome used in this analysis was obtained from the National Center for Biotechnology Information (NCBI) Virus
Variation Database (www.ncbi.nlm.nih.gov). The searching of genome sequence was done through the database using Zika Virus and protein envelopes of about 500 sequence in length.
Multiple sequence alignments. MUSCLE alignment method was used to align the protein sequence downloaded. It was noted that the multiple protein sequence alignments were achieved through the MUSCLE algorithms, using the standard generic translation code, with universal settings, which provided the best results.
Phylogenetic tree. The phylogenetic tree was created from the aligned sequences using PhyML Maximum with probability method utilizing 20 bootstraps replicate searches, which resulted in the best possible scoring tree (Ebranati et al., 2019). The SRT & NNI search were used to perform this tree improvement, resulting in 300 drafts, which complement ZIKV genomes in the NCBI database.
Results

Phylogenetic analysis
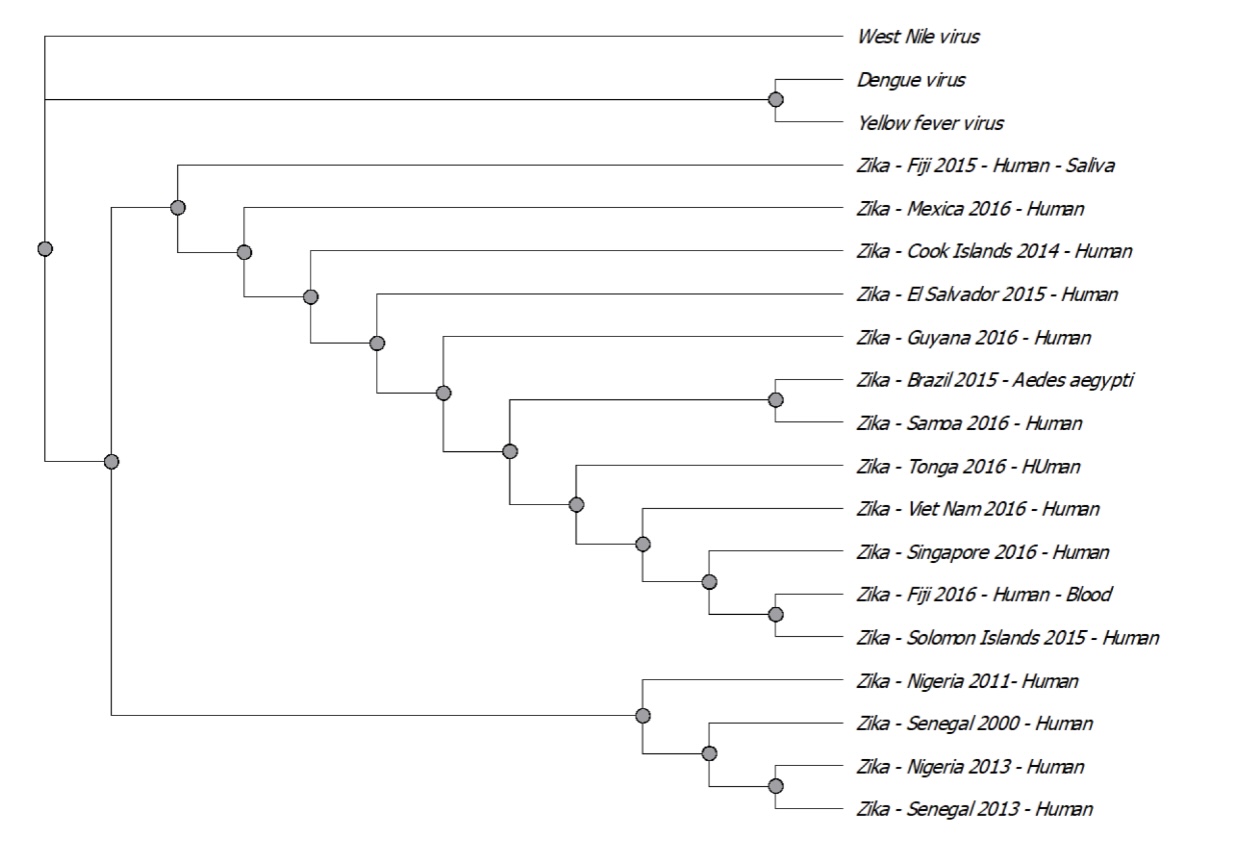
The algorithm used to analyze the maximum likelihood of ZIKV NS5 phylogenetic tree led to the understanding of probabilities of the sequences based on their evolution from one branch to the other.
Discussion and Conclusion
ZIKV is one of the recently emerging pathogen carried by mosquitoes. According to Polonio et al. (2017), it was first analyzed and understood in 1952, when it was separated in 1947 from a sentinel Rhesus macaque and a year later in Aedes Africanus in Ugandan, Zika forest. However, since its first reporting, few cases of infections were registered until 2007, when a massive outbreak happened in Micronesia. According to Musso et al. (2018), it was later detected and confirmed in 2013, when 396 cases were positive from laboratory tests. Ever since, different variants of the virus have continued to affect other parts of the world.
The current and previous phylogenetic tree analyses have mostly identified African and Asian lineages. It has also been understood that the virus is transmitted through mosquitoes, such as those in the Aedes genus, particular in the Culicidae family, when they are in their sylvatic phase. Humans are considered to be the most stable amplification host, where other primates are absent (Scaturro et al., 2018). This indicates that these viruses can survive in primate hosts, but where these are not present, human body becomes the most suitable host for them. Ending the disease has been an issue due to mutation of the virus, changing how the antivirus works from different branches.
The NS5 protein performs different essential functions, which ensure that the virus can replicate and survive, which makes it an important factor to target. While there are several outcomes resulting from the behavior of the virus, it is critical to discuss drug repurposing against the virus. While testing the inhibition behavior for ZIKV NS5, it is possible to screen several drugs against the virus. However, this can consume resources and time, which can be used in other analyses. Consequently, it is critical to examine present nucleoside analogues as these have demonstrated how effective they are in viral inhibition. For instance, these can include the pyrimidine or purine, having similar structure as that of the virus’ RNA or DNA, hence can trick them to grow longer chains, thus terminating earlier than normal. In addition, some other characteristics include the efficiency in uptake of various factors and activating them within the infected cells.
Conclusion
NS5 is critical in the Zika Virus’ survival and infectivity, making it a crucial target for developing an anti-viral drug. Since it has a crystal-like structure, the ZIKV can mimic different groups of antiviruses, thus it is possible to design a drug which can inhibit the activities of RdRp domain or can be made to target multiple allosteric behaviors happening as a result of reaction between NS5 and its various cofactors. Moreover, it is important to repurpose drugs in the present markets, particularly, those which indicate they have potential as anti-NS5 in various flaviviruses as its proteins have conservative nature. It is also crucial to consider the study of the NS5 protein, which can help in understanding the host proteins which need to be targeted to cause maximum ZIKV inactivity.
References
Ebranati, E., Veo, C., Carta, V., Percivalle, E., Rovida, F., Frati, E. R., Amendola, A., Ciccozzi, M., Tanzi, E., Galli, M. and Baldanti, F. (2019). Time-scaled phylogeography of complete Zika virus genomes using discrete and continuous space diffusion models. Infection, Genetics and Evolution, 73(1), 33-43. Web.
Musso, D., Bossin, H., Mallet, H. P., Besnard, M., Broult, J., Baudouin, L., Levi, J. E., Sabino, E. C., Ghawche, F., Lanteri, M.C. and Baud, D. (2018). Zika virus in French Polynesia 2013–14: Anatomy of a completed outbreak. The Lancet Infectious Diseases, 18(5), e172-e182. Web.
Polonio, C. M., de Freitas, C. L., Zanluqui, N. G., & Peron, J. P. S. (2017). Zika virus congenital syndrome: Experimental models and clinical aspects. Journal of Venomous Animals and Toxins including Tropical Diseases, 23(1), 1-9. Web.
Scaturro, P., Stukalov, A., Haas, D.A., Cortese, M., Draganova, K., Płaszczyca, A., Bartenschlager, R., Götz, M. and Pichlmair, A. (2018). An orthogonal proteomic survey uncovers novel Zika virus host factors. Nature, 561(7722), 253-257. Web.
Upadhyay, A. K., Cyr, M., Longenecker, K., Tripathi, R., Sun, C., & Kempf, D. J. (2017). Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta Crystallographica Section F: Structural Biology Communications, 73(3), 116-122. Web.