Cancer is commonly considered to have genetic origins. However, a growing body of evidence suggests that several epigenetic processes (most prominently, histone modification and DNA methylation), have been considered as major contributors to the formation of cancer tumours. Nevertheless, it has remained largely unclear whether changes in epigenetic regulation are associated with the formation of tumours. Recently, a study was published in the Journal of Clinical Investigation, according to which changes in DNA methylation are sufficient for the onset of tumour genesis.
The research team from Baylor College of Medicine used a genetic component expected to attract chemical compounds that suppress genetic functions of cells to trigger the gene silencing process. The said component, known as methyl groups, was introduced into the p16 gene that is normally associated with the control of a cell-division process. The methyl groups used in the study were based on human genome motifs and were previously shown to be associated with the silencing of genes and methylation in the development of humans.
The experiment was done in vivo using mice. The results of the study have shown that 27 percent of mice whose genetic information was modified with the described methylation magnet have developed several types of cancer, including sarcomas, leukaemia, and lung cancer. In addition, 5 percent of the sample that received a copy of a methylation magnet and a copy of wild control modification also developed cancers.
However, no development of tumours was observed in the group receiving a control modification. Finally, both the speed of onset of tumour development and the length of tumour-free survival was adversely impacted by the introduction of the described mutation. The obtained results confirm the validity of a long-standing targeted methylation approach by offering direct functional evidence of a link between epimutation and the formation and malignant progression of tumours.
Epigenetics is a crucial element responsible for the adjustment of genome functionality. In a certain sense, the epigenome can be viewed as a chemical driver that adjusts the ways in which genome primes development of a living organism. The human genome consists of 23 pairs of chromosomes containing information on how the organism should function. The information is stored in portions of DNA referred to as genes. In this regard, a genome is a sum of all information contained in the DNA.
The epigenome, on the other hand, is comprised of chemical compounds that can alter the way in which DNA carry out their functions. On the most basic level, this process can be explained as turning certain DNA segments on and off and, as a result, suppressing or promoting certain functions. It is important to understand that the described process does not introduce permanent or irreversible changes into the genome – rather, it alters functions of cells created using these sets of instructions.
While each individual cell in a human organism contains the entire DNA sequence, the functions and characteristics of these cells may differ significantly across the population. These differences can be traced to the ability of epigenetic compounds to suppress or promote the production of certain proteins. For example, red blood cells are able to fulfil their function due to the development of proteins that deliver oxygen across the human body. In a similar manner, cells in the eye are more suitable for detecting light as a result of the production of proteins with respective functions. While many of the specialised functions of cells are determined by the information contained in a genome, at least some of them can be attributed to the chemical bonds facilitated by epigenome.
The most important distinction between genome and epigenome is the duration of their effect. The composition of genetic information is relatively constant and requires significant time to change. In contrast, the changes created by the attachment of epigenomic chemical compounds are relatively short-lived. In some cases, such modifications emerge within a time span of several generations in response to external factors.
It is important to understand that despite their seemingly temporary nature, the described processes need to be heritable in order to be considered epigenetic changes. The easiest example of epigenetic changes in humans is the occurrence of changes in populations in response to harsh environmental conditions (e.g. drought or famine). It is expected that in response to the scarcity of food, at least some of the subsequent generations will display certain traits and characteristics necessary for successful mitigation of unfavourable conditions.
For instance, it is likely that the children conceived during the period of malnutrition will have decreased bodyweight even if the famine does not occur during their lifetime. In addition, certain changes can be expected in the mental state of the successors – for instance, it is possible to observe lower obesity rates despite equal availability of food, which would be consistent with the restrictions imposed by the expected nutrition deficit. Importantly, the genetic information will be identical in all segments of the population, and all changes will be determined by the suppression of certain cell functions by the attached epigenomic compounds.
As can be seen, the epigenetic changes are largely intended to increase the adaptability of the impacted population to the unfavourable environmental conditions. However, some of the changes may have undesirable consequences. Specifically, the adverse psychological and behavioural disorders that emerge in response to traumatic experience may develop in individuals regardless of the actual exposure to trauma.
As a result, the children who never experienced traumatic events are still at the increased risk of developing mental disorders. It is also important to mention that, according to preliminary data, these changes are reversible. In other words, it is possible to reduce the risks associated with epigenetic effects by modifying the environment accordingly and, as a result, ensure positive long-term effects in the population at large.
Arguably the most significant contribution of epigenetics to the field of healthcare is its role in cancer development. Cancer have long been considered partially determined by genetic code, primary due to the identification of mutations at the early stages of common cancers. However, only a small fraction of the said genetic mutations was positively associated with tumour development, leading some scientists to consider alternative explanations. Currently, it is commonly agreed that epigenetic changes are at least equally important in the development of tumours as genetic mutations.
For example, the occurrence of cellular transformation results from a mutation in the DNA, whereas the probability of metastasising increases as a result of the changes in epigenome. In simple terms, genetic changes are necessary to make the development of cancer possible, but unless the epigenetic changes are involved, this possibility is negligible from the practical perspective. Next, genetic changes of the epigenetic enzyme-encoding process may produce changes in the epigenome and, by extension, contribute to cancer development through the activation of an oncogene. The process of binding of proteins to epigenetic marks can also be impacted by alterations in the genetic code, further strengthening the connection between genetic and epigenetic contributors to the disease.
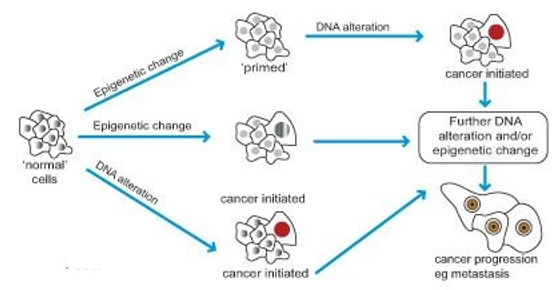
One of the most prominent examples of the epigenetic alteration that was considered responsible for the incidence of cancer is DNA methylation. In the simplest terms, it can be described as the process in which tumour suppressing genes get switched off by epigenetic process while overall stability of the genome decreases and some oncogenes are activated. Thus, over-expression of DNA methylation enzyme in cells is associated with a higher likelihood of cancer development in humans.
However, it should be understood that despite the apparent risk, the said enzymes play an important role in cell functioning and are thus crucial for adequate functioning of an organism. In addition, DNA methylation seems to be closely associated with histone methylation, another important element of epigenetic mechanism. In fact, it highly probable that the two processes support and reinforce each other, and introducing changes to one of them creates severe imbalance and disrupts epigenetic changes on a large scale.
Several factors are known to contribute to DNA methylation, thus increasing the probability of epigenetic cause of cancer. The first such factor is the ageing process. In some tissues, the onset of ageing causes an overall decrease in methylation combined with increased methylation in some areas, which creates conditions characteristic for cancer cells. It is possible to consider that the described change is responsible for the increased occurrence of cancers among elderly people.
Second, some of the components necessary for a functional methylation process, such as methionine and folate, cannot be synthesised and are usually obtained from nutrition. As a result, the deficit of these compounds in the human diet leads to disruption of the DNA methylation process, and, by extension, increases the likelihood of cancers. Third, some of the chemical compounds are able to affect the methylation process. For instance, cadmium is known to inhibit the overall methylation process, and arsenic has been shown to decrease methylation of a specific gene. Therefore, the organisms exposed to these chemical agents in the environment are at a greater risk of developing cancers.
Considering the information above, it would be reasonable to suggest that controlling the methylation process provides humanity with an opportunity to manage and treat cancers. Nevertheless, the described area of research is still in the relatively early stage of development. While numerous studies exist that document observations consistent with the suggested relationship, more reliable data is necessary to conclusively establish it before the knowledge can be implemented in practice.
The study by Yu et al. takes this one step further by isolating a specific gene responsible for the suppression of tumour development and silencing it. As a result, it becomes possible to determine whether DNA methylation has the hypothesised effect on cancer development and, if it does, what is the extent of its influence compared to other factors (e.g. compared to the disruptive effect of increased total methylation). As was mentioned above, the study results indicate the validity of these suggestions by establishing a direct connection between the silencing of a tumour-suppressing gene and the formation of cancer.
However, the causal relationship between the two has not been conclusively established. In this regard, the study in question serves as proof that at least some epigenetic changes are responsible for the development of cancer, which adds to our current understanding of the condition and outlines directions for further research. Another and, perhaps, more important implication is the identification of treatment opportunities for cancers.
Currently, epigenetic changes have been positively identified with most cancers, which suggests that the obtained results are applicable to a large number of cases. Once it is conclusively proven that silencing the tumour-suppressing gene results in the increased chance of developing cancer, it is possible to imply that an instrument can be developed that would detect similar epigenetic scenarios, leading to more accurate diagnoses and, by extension, higher efficiency of treatment. It is also possible to expect that once sufficient evidence is accumulated, new treatment options can be developed that utilise epigenetic mechanisms. For instance, reactivating the tumour-suppressing gene may decrease the likelihood of cancer development, as was demonstrated by the outcomes in the control group.
Epigenetic mechanisms play an important role in the onset and development of cancer. Despite significant progress made in the field, the research in the area is still in its early phase. The results demonstrated by the study enhance our theoretical knowledge on the matter and outline a viable direction in epigenetic engineering that may offer improvements to cancer diagnosis and treatment process.
References
- Schübeler, D. Nature 517, 321-326 (2015).
- Yu, et al. J. Clin. Inv. 124, 3708-3712 (2014).
- Shen, L., et al. PLoS Gen. 3, 2023-2036 (2007).
- Feil, R., & Fraga, M. F. Nature Rev. Gen. 13, 97-109 (2012).
- Szyf, M. Trends Mol. Med. 21, 134-144 (2015).
- Gapp, K., et al. Neuropsychopharmacology 41, 2749-2758 (2016).
- Dawson, M. A., & Kouzarides, T. Cell 150, 12-27 (2012).
- Bannister, A. Abcam. (n.d.). Web.
- Rasmussen, K. D., & Helin, K. Gen. & Dev. 30, 733-750 (2016).
- Khan, S., Shukla, S., Sinha, S., & Meeran, S. M. Exp. Op. Therap. Targ. 20, 689-703 (2016).