Introduction
Most pyruvate would accumulate in yeast suspension 1 (1g yeast in 10cm3 Na2HPO4). The optimum pH for pyruvate decarboxylase, an enzyme that converts pyruvate to ethanal, is about 4.3-6.7. Na2HPO4 raises the pH to 8-11 (weakly basic), which inactivates the enzyme (Garrett and Grisham, 2016). In contrast, KH2PO4 lowers the pH to 4.4-4.7, which would favour pyruvate decarboxylation to produce ethanal. Hence, more pyruvate would accumulate in solution 1 than in solution 2.
Colours and intensity
Solution 1 has the most intense colour.
Tube with most pyruvate
Tube 1
Agreement with the prediction
Yes. The red colour observed in tube 1 during the 2,4-dinitrophenylhydrazine test indicates a significant amount of pyruvate. In contrast, tube 2 produced a moderate amount of pyruvate consistent with the pale orange colour observed. Pyruvate decarboxylase is repressed in weakly basic conditions (tube 1); therefore, pyruvate accumulates and is detected by 2,4-dini-trophenylhydrazine.
If yeast is grown in the presence of sodium sulphite, ethanal (acetaldehyde) accumulates, and its presence can be detected. However, under slightly alkaline conditions in the presence of sodium sulphite, pyruvate accumulates but ethanal does not. Deduce, explaining your reasoning.
The sequence metabolites
Sodium sulphite, a trapping agent, forms adducts with ethanal that can be detected chemically. In yeast, phosphoenol pyruvate is converted to pyruvate by pyruvate kinase. Subsequently, pyruvate decarboxylase converts the pyruvate, a glycolytic product, into ethanal and releases CO2. Ethanal is then reduced to ethanol and NaD+ by a dehydrogenase enzyme as shown below.
The results of muscle tissue
In a muscle tissue, pyruvate would be converted to lactate instead of ethanal. Lactate dehydrogenase converts pyruvate to lactic acid and oxidizes NaDH to NaD+ as shown below.
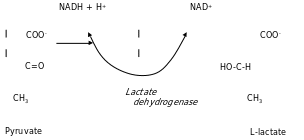
Practical II: The NAD Requirements of Lactate Dehydrogenase
Prediction of lactate formation
It is predicted that most pyruvate in Test 2 and 3 would be converted to lactate. The two incubation mixtures contain the NADH and NADPH cofactors for the LDH oxidation of pyruvate to lactate.
Results
Table 1
- Record the absorbance readings in the table above
Calculations of the amount of pyruvate converted
Amount of pyruvate converted is given by:
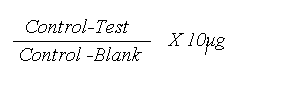
- Tube 1 = 2.243-1.915/2.243-0.986 x 10 μg = 2.6 μg
- Tube 2 = 2.243-1.915/2.243-0.986 x 10 μg = 2.6 μg
- Tube 3 = 2.243-2.015/2.243-0.986 x 10 μg = 1.814 μg
- Tube 4 = 2.243-0.986/2.243-0.986 x 10 μg = 10 μg
- Tube 5 = 2.243-2.243/2.243-0.986 x 10 μg = 0.0 μg
Drawn conclusions
The incubation mixtures 1 and 2 have the most enzyme (LDH) activity compared to other tubes. The amount of pyruvate converted to lactate in these tubes 1 and 2 is the highest (2.6 μg). LDH requires NADH to convert pyruvate to lactate (Garrett and Grisham, 2016). The phosphate buffer (tube 1) and NADH (tube 2) enhance LDH activity. NAPDH, an equivalent of NADH, has a less effect on LDH activity.
Does this fit with your prediction?
The findings partly fit with the predicted results. The highest amount of pyruvate converted to lactate is seen in tube 1, which is consistent with the prediction.
Practical III: Hydrolysis of Organic Phosphate
Briefly giving reasons, predict whether (1) AMP, (2) ADP, (3) ATP, (4) glucose 6-phosphate will be hydrolysed by hot acid
- AMP – the molecule has a single phosphate molecule, and thus, the hot acid will hydrolyse it to adenosine and Pi.
- ADP – hot acid can hydrolyse ADP to adenosine monophosphate (AMP), inorganic phosphate (Pi), and energy due to the high energy phosphoanhydride bond.
- ATP – the molecule will be hydrolysed to ADP and Pi for it is a high charge density due to the three phosphate units in ATP makes the molecule unstable (Garrett and Grisham, 2016). Thus, the hot acid will easily hydrolyse ATP in an exergonic process.
- Glucose 6-phosphate – G 6-P is less labile because it is an ester phosphate. Therefore, hot acid will not hydrolyse this molecule.
Tabulation of absorbance data table below
Table 1
Blank = 0.00
Completion of the table below (80μg phosphate = 0.84μmole)
With the standard curve of phosphate molybdate of 0.25nm/1ug, the table was filled as follows:
Determination of micro-grams
AMP Calculations
0.25nm has 1µg, what about 0.050nm at 0oC and 0.057nm at 1000oC
- Absorbance of 0.050nm at 0oC has 1/0.25nm X 0.05 = 0.2 µg
- Absorbance of 0.057 at 1000oC has 1/0.25 X 0.057 = 0.228 µg
ADP Calculations
- Absorbance of 0.082nm at 0oC has 1/0.25nm X 0.082 = 0.328 µg
- Absorbance of 0.082nm at 100oC has 1/0.25nm X 0.034 = 0.136 µg
ATP Calculations
- Absorbance of 0.082nm at 0oC has 1/0.25nm X 0.244 = 0.976 µg
- Absorbance of 0.082nm at 100oC has 1/0.25nm X 0.034 = 0.136 µg
G-6-P Calculations
- Absorbance of 0.082nm at 0oC has 1/0.25nm X 0.040 = 0.16 µg
- Absorbance of 0.082nm at 100oC has 1/0.25nm X 0.041 = 0.164 µg
Determination of micro-moles
Since 80μg phosphate = 0.84μmole), 1µg has (0.84mole/80µg) = 0.0105 moles
AMP Calculations
- At 0oC, 0.2 µg has 0.2 X 0.0105 = 0.0021 moles
- At 100oC, 0.228 µg has 0.228 X 0.0105 = 0.0024 moles
ADP Calculations
- At 0oC, 0.328 µg has 0.328 X 0.0105 = 0.0034 moles
- At 100oC, 0.136 µg has 0.136 X 0.0105 = 0.0014 moles
ADP Calculations
- At 0oC, 0.976 µg has 0.976 X 0.0105 = 0.0102 moles
- At 100oC, 0.136 µg has 0.136 X 0.0105 = 0.0014 moles
G-6-P Calculations
- At 0oC, 0.16 µg has 0.16 X 0.0105 = 0.0017 moles
- At 100oC, 0.164 µg has 0.164 X 0.0105 = 0.0017 moles
Table 2
Stability of phosphates at 100oC
AMP and G-6-P are stable phosphates at 100oC because their absorbance, micrograms, or moles remain relatively constant irrespective of the temperature change from 0oC to 100oC.
Does this agree with your prediction?
The outcome does not agree with my prediction for I assumed that heat catalyzes the hydrolysis of all organic phosphates.
The extent of phosphate hydrolysis
- In AMP, hot acid increases hydrolysis (0.0024 – 0.0021) 0.0003 moles
- In ADP, hot acid reduces hydrolysis by (0.0034 – 0.0014) 0.002 moles
- In ATP, hot acid reduces hydrolysis by (0.0102 – 0.0.0014) 0.0088 moles
- In G-6-P, hot acid does not influence hydrolysis for equal moles of phosphate were hydrolyzed.
Practical IV: The Phosphorylation of Creatine by Creatine Kinase
Prediction
Based on free energy changes, I expect ADP and ATP to be suitable phosphate donors in the biochemical reaction that synthesises creatine phospahate because they have high free energies (-37), which are equal to that of their product (-37).
Tabulation of results
Blank = 0.000
Tabulation of calculations
Using the standard curve of phosphate molybdate of 0.25nm/1ug, the table was filled as follows:
G-6-P Calculations
= 0.025 nm/0.25 nm X 1 µg= 0.1 µg
AMP Calculations
= 0.025 nm/0.25nm nm X 1 µg = 0.1 µg
ADP Calculations
=0.025 nm/0.25 nm nm X 1 µg= 0.1 µg
ATP Calculations
= 0.019 nm /0.25nm nm X 1 µg= 0.076 µg
Buffer Calculations
= 0.043 nm/0.25 nm X 1 µg= 0.172 µ
Calculation of Moles
- Since 80μg phosphate = 0.84μmole), 1µg has (0.84mole/80µg) = 0.0105 moles
- G-6-P moles = 0.0105 X 0.1 = 0.00105 moles
- AMP moles = 0.0105 X 0.1 = 0.00105 moles
- ADP moles = 0.0105 X 0.1 = 0.00105 moles
- AMP moles = 0.0105 X 0.076 = 0.000798 moles
- Buffer moles = 0.0105 X 0.172 = 0.0018 moles
Phosphate donors
- In the experiment, ADP and ATP act as phosphate because they have two and three phosphates in their structure, which they can easily donate for they have high free energy when compared to other phosphates.
- G-6-P, AMP, and buffer act do not act as phosphate donors because they have single phosphate molecules in their structure and low free energy.
Agreement of results
Results do not agree with the prediction because it shows that all organic phosphates, namely, G-6-P, AMP, ADP, ATP, and buffer were phosphate donors in the experiment.
Brief account
G-6-P, AMP, and ADP are phosphates that donated equal amounts of phosphates in the formation of creatine phosphates because each donated 0.00105 moles. ATP donated the lowest moles of phosphates (0.000798) while buffer donated the highest moles of phosphates (0.0018).
Bibliography
Garrett, H. and Grisham, M. (2016) Biochemistry, 15th edn., Boston: Harcourt Inc.