Introduction
Taxol is a plant solution used in treatment of ovarian and breast cancer. The solution is found in the bark of the Pacific yew tree. Taxol is highly used in medical industry for chemotherapy treatment on ovarian and breast cancer victims. In medical terms, ovarian cancerous cells referred to as SKOV3 cells while breast cancer cells are known as MCF7 cells. Taxol kills cancerous cells through a mechanism known as microtube depolymerisation. This mechanism entails changing of the microtubule dynamics of the cells. Such situation results to destruction of the microtube structure of the cancer cells. All cancer cells require the microtube structure for growth and mitosis. Consequently, microtube destruction leads to death of most cancerous cells (Chiang 2012, p. 34).
Today, Taxol is used in treatment of ovarian and breast malignant growths. It works in a similar wy as the taxanes anticancer agents hence health experts have categorized it in taxanes class. Naturally, cancer cells grow continuously and they reproduce through dividing to yield more cells and grow in size. Each new cell grows uncontrollably with time leading to a malignant growth in various body parts. Because Taxol inhibits further multiplication and growth of cancer cells, it prevents them from spreading to other body parts especially in ovaries and breast (Wu 2012, p. 32). Currently, research is underway to examine if Taxol can treat other malignant growth on other body parts (Chiang 2012, p. 53). However, the first time Taxol was introduced for cancer treatment, critics argued that it had side effects on women body at long run (Goodman & Walsh 2001).
Aims of the experiment
The following were the objectives of this experiment:
- To determine the effectiveness of Taxol in inhibiting breast cancer cells (MCF7) and ovarian cancer cells (SKOV3) using culture method.
- To study advanced cell examination methods that do not involve staining the cells. They included how to:
- Carry out Vitro culture cell harvesting using trypsin enzymes.
- Count cell using haemocytometer.
- Determine cell viability with help of trypan blue reagent.
- Dilute cell suspensions to the expected concentration for use in experimental procedures.
- Plot a standard curve for use in cell counting assay.
- Carry out cytotoxicity assay experiments and analyse the results
Methods
Procedures
Protocol 1: Observing live cells: Since we desired to observe the cells without killing them, we used differential interference, dark field and, phase-contrast to observe the cell as described in the below table.
Protocol 2: Harvesting adherent cells from the culture
Protocol 3: using trypan blue to carry out vital staining. We blended trypan blue stain with 100 of cell suspension in a microfuge tube. Using haemocytometer, we determined the number of dead and live cancer cells.
Protocol 4: Counting cells with the haemocytometer: We placed the cover slip on the counting chamber of haemocytometer using microscope. We put 10 µL of the suspension in the cover slip cover slip and countered the viable cells and dead cells in the corner square and the middle square and recoded the cells in table 1.1. We calculate number of cells per ml in the suspension. We further calculated the cell viability from data obtained. We also calculated the amount of cell suspension required to prepare ml of culture containing 25000 viable cells.
Protocol 5: preparation of viable cell standards. We prepared 10 microfuge tubes using 200µL of RPI-FBS culture. We then diluted the original suspension using pipette. We divided the suspension into 12 test tubes and labeled them A1-A11.
Protocol 6: tetrazolium sal-based cell counting assay. We prepared a suspension by adding 100µL to tubes A1-A12. We then added RPMI/FBS to A12 only. We further added 100µL of the MCF& to B2-B12. RPMI/FBS to A12 was added to B12 only. The demonstrator added CCK-8 to the experiment. We incubated the solution for 1 hour at a temperature of 37°C. We finally read the absorbance at 490 nm and recorded the results.
Results
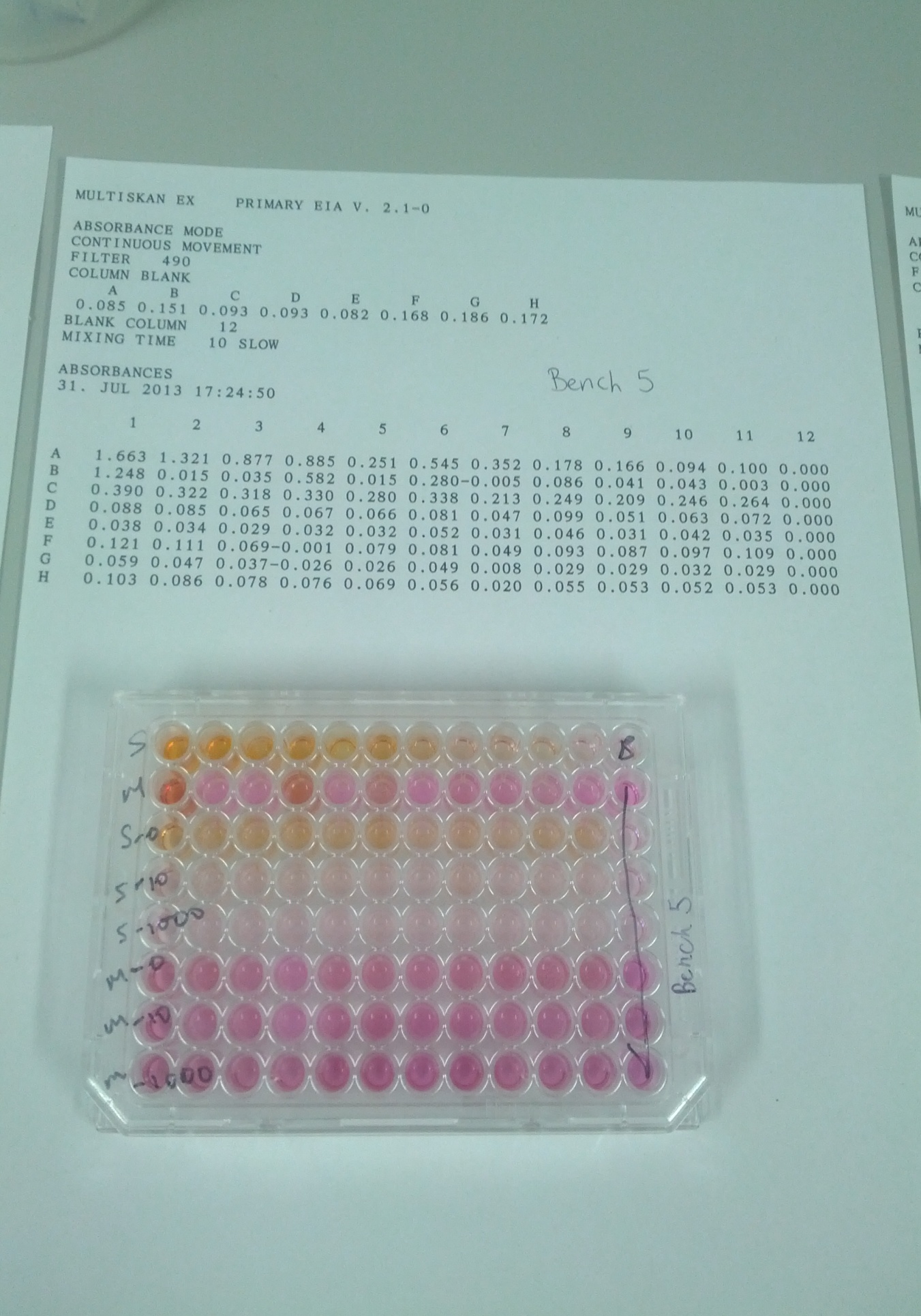
The following table shows the relationship between absorbance and the corresponding number of cells found inside the well at the end of the experiment.
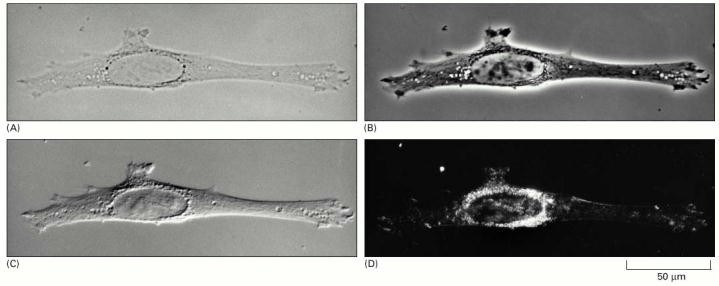
Figure 2 above shows the four types of light microscopy. Above is a photo of 4 fibroblast cell inside the culture. (A) Bright-field microscopy. (B) Phase-contrast microscopy. (C) Nomarski differential- interference-contrast (DIC) microscopy. (D) Dark-field microscopy (Alberts, Johnson & Lewis 2002, P. 40).
Calculation of cell viability
Cell viability is given by:
No. of Viable Cells Counted x 100 = % viable cells Total no. cells counted (viable + dead)
- The number of cells in 1ml suspension was= 1.96×10^5 cells.
- The number of viable cells was = 8.18×10^4 cells/ml
- How much of your cell suspension would you need to prepare 1mL of culture that has 25,000 viable cells per ml? Use the equation C1V1= C2V2
C1V1=C2V2
V1= 25,000×1÷8.18×10^4 = 30562347.19
MCF7 SKOV3
T=total 1.96×10^5 T= 1.84×10^6
L=Live 8.18×10^4 so 42% cells/ml L= 1.83×10^6 So 100% cells/ml
Protocol 8: Data analysis
Standard curve data
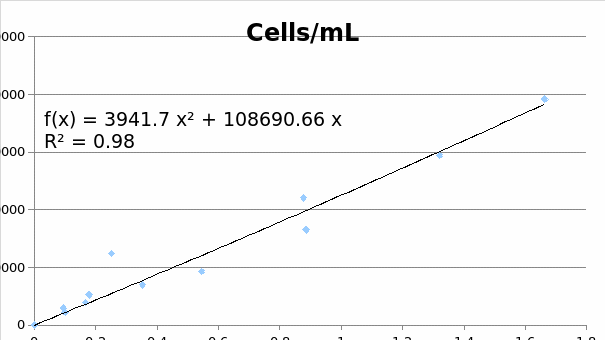
MCF7
SKOV3
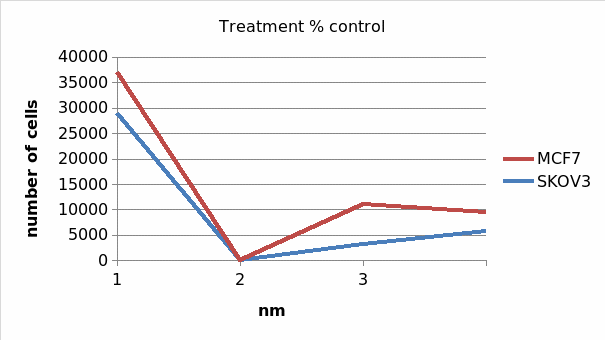
Table 1.3: Average number of viable cancer cells after 24hrs
Treatment Average number of cells
Table 1.4 treatment percentage control
Discussion
Answering questions
2 (a) Yes, there are fewer cells in taxol-treated controls than in non-treated controls. From table 1.3, SKOV3 onm control had 28921 alive cancer cells compared to 7121 in 10nm and 3646 in 1000nm. Similarly, MCF 0nm taxol treated control had 8134 live cells compared to 2894 in 10nm and 6366 in 1000nm. Therefore, it is true that taxol treated controls had fewer cells than in onm taxol treated controls.
(b) Yes, MCF 1000nm taxol control had 6366 alive cancer cells after 24 hours from a total of 8134 at 0nm. On the other side, SKOV3 1000nm taxol control had a total of 3646 alive cancer cells with onm control having 28921 alive cells. This implies that, taxol is more poisomous on SKOV3 (Ovarian cancer cells) than it is on MCF7 (breast cancer cells).
5. No, this is due to the fact that taxol treated control of MCF7 and SKOV3 do not process Caspase-9. However, the same treatment can activate caspase-3. Generally, when caspase-3 is not well induced in the presence of SKOV3 cells, caspase-9 cells are not activated. This is the reason why the results do not match.
The above results reveal that Taxol is more effective in treatment of breast cancer than it does on the ovarian cancer. This is evident from the fact that at the end of the experiment, the number of live cells in SKOV3 was 100%. The corresponding propotion in MCF7 was 42%. This implied that 58% of the breast cancer cells died on the exposure to the Taxol agent in MCF7. In addition, research asserts that Taxol agent has the same effects on the two cells (Balzer 2010, p. 67). The result further implies that none or few SKOV3 cells died on exposure to the Taxol cancer treating agent. Therefore, we recommend that Taxol should be highly used in treatment of breast cancer (Wu 2012, p. 78).
From figure 2, it is clear that the amount of brightness influence the visibility of the cancer cells. The figure further shows that phase contrast microscope brightness yields the best view of the cancer cells. In contrast, dark field microscopy produces the darkest image, which is difficult for the examiner to identify various components making up the cell. DIC microscopy is moderately bright while bright field microscopy produces a very bright image. We therefore recommend that, phase contrast microscope be used for Taxol cancer examination experiments.
References
Alberts, B, Johnson, A, & Lewis, J 2002, Looking at the Structure of Cells in the Microscope, Molecular Biology of the Cell, Garland Science, New York.
Balzer, M 2010, Molecular Alterations that Reduce Cortical Containment of Tubulin Microtentacles and their Impact on Breast Tumor Metastasis, Defense Technical Information Center, Ft. Belvoir.
Chiang, Y 2012, Overexpression of [alpha]-catulin promotes taxol resistance in ovarian cancer, Cambridge University Press, Cambridge.
Goodman, J & Walsh, V 2001, The story of taxol: nature and politics in the pursuit of an anti-cancer drug, Cambridge University Press, Cambridge.
Wu, W 2012, Genome-Free Viral Capsids for Targeted Drug Delivery to Breast Cancer, Cambridge University Press, Cambridge.